Abstract
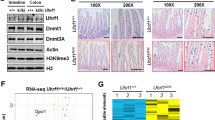
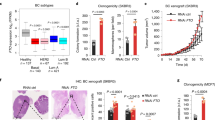
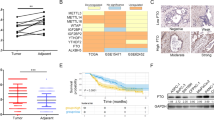
PRDM proteins are tissue-specific transcription factors often deregulated in diseases, particularly in cancer where different members have been found to act as oncogenes or tumor suppressors. PRDM5 is a poorly characterized member of the PRDM family for which several studies have reported a high frequency of promoter hypermethylation in cancer types of gastrointestinal origin. We report here the characterization of Prdm5 knockout mice in the context of intestinal carcinogenesis. We demonstrate that loss of Prdm5 increases the number of adenomas throughout the murine small intestine on an ApcMin background. By using the genome-wide ChIP-seq (chromatin immunoprecipitation (ChIP) followed by DNA sequencing) and transcriptome analyses we identify loci encoding proteins involved in metabolic processes as prominent PRDM5 targets and characterize monoacylglycerol lipase (Mgll) as a direct PRDM5 target in human colon cancer cells and in Prdm5 mutant mouse intestines. Moreover, we report the downregulation of PRDM5 protein expression in human colon neoplastic lesions. In summary, our data provide the first causal link between Prdm5 loss and intestinal carcinogenesis, and uncover an extensive and novel PRDM5 target repertoire likely facilitating the tumor-suppressive functions of PRDM5.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fog CK, Galli GG, Lund AH . PRDM proteins: important players in differentiation and disease. Bioessays 2012; 34: 50–60.
Duan Z, Person RE, Lee HH, Huang S, Donadieu J, Badolato R et al. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol 2007; 27: 6889–6902.
Burkitt Wright EM, Spencer HL, Daly SB, Manson FD, Zeef LA, Urquhart J et al. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am J Hum Genet 2011; 88: 767–777.
Cheng HY, Chen XW, Cheng L, Liu YD, Lou G . DNA methylation and carcinogenesis of PRDM5 in cervical cancer. J Cancer Res Clin Oncol 2010; 136: 1821–1825.
Deng Q, Huang S . PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene 2004; 23: 4903–4910.
Shu XS, Geng H, Li L, Ying J, Ma C, Wang Y et al. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/beta-catenin signaling and is frequently silenced in multiple tumors. PLoS One 2011; 6: e27346.
Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 2009; 136: 2149–2158.
Watanabe Y, Toyota M, Kondo Y, Suzuki H, Imai T, Ohe-Toyota M et al. PRDM5 identified as a target of epigenetic silencing in colorectal and gastric cancer. Clin Cancer Res 2007; 13: 4786–4794.
Galli GG, Honnens de Lichtenberg K, Carrara M, Hans W, Wuelling M, Mentz B et al. Prdm5 regulates collagen gene transcription by association with RNA polymerase II in developing bone. PLoS Genet 2012; 8: e1002711.
Meani N, Pezzimenti F, Deflorian G, Mione M, Alcalay M . The tumor suppressor PRDM5 regulates Wnt signaling at early stages of zebrafish development. PLoS One 2009; 4: e4273.
Reya T, Clevers H . Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–850.
Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991; 66: 589–600.
Kinzler KW, Vogelstein B . Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–170.
Clevers H . Wnt/beta-catenin signaling in development and disease. Cell 2006; 127: 469–480.
Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 2004; 18: 1385–1390.
Nathke IS . The adenomatous polyposis coli protein: the Achilles heel of the gut epithelium. Annu Rev Cell Dev Biol 2004; 20: 337–366.
Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene 1994; 9: 603–609.
Moser AR, Pitot HC, Dove WF . A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990; 247: 322–324.
Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF . Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 2010; 140: 49–61.
Ye L, Zhang B, Seviour EG, Tao KX, Liu XH, Ling Y et al. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett 2011; 307: 6–17.
Hidalgo IJ, Raub TJ, Borchardt RT . Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989; 96: 736–749.
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB . The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7: 11–20.
Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB . Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 2008; 18: 54–61.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Menendez JA, Lupu R . Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 2007; 7: 763–777.
Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol 2011; 18: 846–856.
Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB . ATP citrate lyase is an important component of cell growth and transformation. Oncogene 2005; 24: 6314–6322.
Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 2005; 8: 311–321.
Kim D, Salzberg SL . TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 2011; 12: R72.
Roberts A, Pimentel H, Trapnell C, Pachter L . Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011; 27: 2325–2329.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3.
Sanges R, Cordero F, Calogero RA . oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics 2007; 23: 3406–3408.
Acknowledgements
We would like to acknowledge Julia Sidenius Johansen for her advice on TMA stainings, Bettina Mentz for excellent technical assistance and Fengqin Jia for mouse genotyping. This work was supported by the Danish National Research Foundation, the Danish National Advanced Technology Foundation, the Novo Nordisk Foundation, the EC FP7 programs (ONCOMIRS, grant agreement number 201102), the Lundbeck Foundation and the Danish Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Disclaimer
This publication reflects only the authors’ views. The commission is not liable for any use that may be made of the information herein.
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Galli, G., Multhaupt, H., Carrara, M. et al. Prdm5 suppresses ApcMin-driven intestinal adenomas and regulates monoacylglycerol lipase expression. Oncogene 33, 3342–3350 (2014). https://doi.org/10.1038/onc.2013.283
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.283
Keywords
This article is cited by
-
Tumor-suppressive functions of protein lysine methyltransferases
Experimental & Molecular Medicine (2023)
-
Low expression of PRDM5 predicts poor prognosis of esophageal squamous cell carcinoma
BMC Cancer (2022)
-
PRDM1/BLIMP1 induces cancer immune evasion by modulating the USP22-SPI1-PD-L1 axis in hepatocellular carcinoma cells
Nature Communications (2022)
-
Research progress on FASN and MGLL in the regulation of abnormal lipid metabolism and the relationship between tumor invasion and metastasis
Frontiers of Medicine (2021)
-
Endocannabinoid signaling regulates the reinforcing and psychostimulant effects of ketamine in mice
Nature Communications (2020)