Abstract
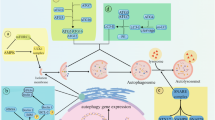
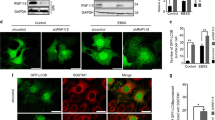
Autophagy is a self-digesting mechanism responsible for the degradation and recycling of most intracellular macromolecules and the removal of damaged organelles by the lysosome. An impressive number of recent studies have provided key information about the regulation of autophagy and its role in cell survival during nutrient depletion and many other stressful situations. In particular, many evidences have highlighted a crucial role of dysregulated autophagy in oncogenesis. Perturbations of the autophagic pathway have been shown to contribute to tumor development. Moreover, cancer cells have developed several mechanisms that allow them to evade chemotherapy-induced cell death, as well as to use autophagy-associated pathways, to potentiate their survival. In this regard, a complex crosstalk between autophagy and apoptosis has recently emerged; the understanding of the molecular mechanisms regulating this interplay may provide new hints on how to properly modulate these processes to halt cancer. Indeed, key proteins originally thought to be apoptosis-specific inhibitors also block autophagy, while apoptosis proteolytic enzymes hamper autophagy by cleaving autophagy-specific proteins and, in some cases, converting them into proapoptotic factors. This review is focused on the role that Ambra1, a central component of the autophagosome formation machinery, has in the switch between autophagy and apoptosis and its implication in cancer development and chemotherapy resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klionsky DJ . Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8: 931–937.
Mizushima N . Physiological functions of autophagy. Curr Top Microbiol Immunol 2009; 335: 71–84.
Deretic V, Levine B . Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5: 527–549.
Kroemer G, Marino G, Levine B . Autophagy and the integrated stress response. Mol Cell 2010; 40: 280–293.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075.
Mizushima N, Yoshimori T, Ohsumi Y . The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27: 107–132.
Orenstein SJ, Cuervo AM . Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol 2010; 21: 719–726.
Burman C, Ktistakis NT . Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett 2010; 584: 1302–1312.
Mehrpour M, Esclatine A, Beau I, Codogno P . Overview of macroautophagy regulation in mammalian cells. Cell Res 2010; 20: 748–762.
Jahreiss L, Menzies FM, Rubinsztein DC . The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008; 9: 574–587.
Dikic I, Johansen T, Kirkin V . Selective autophagy in cancer development and therapy. Cancer Res 2010; 70: 3431–3434.
Li WW, Li J, Bao JK . Microautophagy: lesser-known self-eating. Cell Mol Life Sci 2012; 69: 1125–1136.
Arias E, Cuervo AM . Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 2011; 23: 184–189.
Yang Z, Klionsky DJ . Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22: 124–131.
Jung CH, Ro SH, Cao J, Otto NM, Kim DH . mTOR regulation of autophagy. FEBS Lett 2010; 584: 1287–1295.
Laplante M, Sabatini DM . mTOR signaling in growth control and disease. Cell 2012; 149: 274–293.
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20: 1981–1991.
Simonsen A, Tooze SA . Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186: 773–782.
Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182: 685–701.
He C, Levine B . The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22: 140–149.
Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447: 1121–1125.
Itakura E, Kishi C, Inoue K, Mizushima N . Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19: 5360–5372.
Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q . Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008; 105: 19211–19216.
Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11: 385–396.
Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT . Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11: 468–476.
Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998; 72: 8586–8596.
Sinha S, Levine B . The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 2008; 27 (Suppl 1): S137–S148.
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122: 927–939.
Wei Y, Pattingre S, Sinha S, Bassik M, Levine B . JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30: 678–688.
Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M et al. Beclin1: a role in membrane dynamics and beyond. Autophagy 2012; 8: 6–17.
Abrahamsen H, Stenmark H, Platta HW . Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett 2012; 586: 1584–1591.
Kang R, Zeh HJ, Lotze MT, Tang D . The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571–580.
Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J 2011; 30: 1195–1208.
Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191: 155–168.
Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell 2012; 47: 359–370.
Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141: 656–667.
Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci 2011; 31: 10249–10261.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752.
Fimia GM, Piacentini M . Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci 2010; 67: 1581–1588.
Chang NC, Nguyen M, Germain M, Shore GC . Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J 2010; 29: 606–618.
Levine B, Sinha S, Kroemer G . Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 2008; 4: 600–606.
Adams JM, Cory S . Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol 2007; 19: 488–496.
Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11: 1355–1362.
Wei Y, Sinha S, Levine B . Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008; 4: 949–951.
Lei K, Davis RJ . JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA 2003; 100: 2432–2437.
Donovan N, Becker EB, Konishi Y, Bonni A . JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J Biol Chem 2002; 277: 40944–40949.
Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC . Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett 2009; 274: 95–100.
Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1: e18.
Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW et al. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res 2011; 71: 3625–3634.
Luo S, Rubinsztein DC . Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17: 268–277.
Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis 2011; 2: e144.
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8: 1124–1132.
Betin VM, Lane JD . Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 2009; 122: 2554–2566.
Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010; 142: 590–600.
Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A . The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell 2011; 44: 698–709.
Hou W, Han J, Lu C, Goldstein LA, Rabinowich H . Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 2010; 6: 891–900.
Cecconi F, Piacentini M, Fimia GM . The involvement of cell death and survival in neural tube defects: a distinct role for apoptosis and autophagy? Cell Death Differ 2008; 15: 1170–1177.
Corazzari M, Fimia GM, Piacentini M . Dismantling the autophagic arsenal when it is time to die: concerted AMBRA1 degradation by caspases and calpains. Autophagy 2012; 8: 1255–1257.
Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ 2012; 19: 1495–1504.
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402: 672–676.
Mathew R, White E . Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev 2011; 21: 113–119.
Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25: 795–800.
Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–1075.
Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 2011; 193: 275–284.
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64.
Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M . A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis 2010; 1: e40.
Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59: 59–65.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112: 1809–1820.
Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8: 688–699.
Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9: 1142–1151.
Amaravadi RK, Thompson CB . The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res 2007; 13: 7271–7279.
Livesey KM, Tang D, Zeh HJ, Lotze MT . Autophagy inhibition in combination cancer treatment. Curr Opin Investig Drugs 2009; 10: 1269–1279.
Armstrong JL, Corazzari M, Martin S, Pagliarini V, Falasca L, Hill DS et al. Oncogenic B-RAF signaling in melanoma impairs the therapeutic advantage of autophagy inhibition. Clin Cancer Res 2011; 17: 2216–2226.
Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 2009; 119: 1359–1372.
Velasco G, Sanchez C, Guzman M . Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer 2012; 12: 436–444.
Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011; 334: 1573–1577.
Behrends C, Sowa ME, Gygi SP, Harper JW . Network organization of the human autophagy system. Nature 2010; 466: 68–76.
Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J et al. Function and molecular mechanism of acetylation in autophagy regulation. Science 2012; 336: 474–477.
Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 2008; 105: 10762–10767.
Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 2008; 31: 438–448.
Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011; 44: 325–340.
Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 2011; 10: 013284.
Bennett EJ, Rush J, Gygi SP, Harper JW . Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 2010; 143: 951–965.
Jin J, Arias EE, Chen J, Harper JW, Walter JC . A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23: 709–721.
Schwartz AL, Ciechanover A . Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 2009; 49: 73–96.
Hoeller D, Dikic I . Targeting the ubiquitin system in cancer therapy. Nature 2009; 458: 438–444.
Lipkowitz S, Weissman AM . RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011; 11: 629–643.
UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 2012; 40 (Database issue): D71–D75.
Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012; 40 (Database issue): D261–D270.
Maglott D, Ostell J, Pruitt KD, Tatusova T . Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 2011; 39 (Database issue): D52–D57.
Acknowledgements
This work was supported by grants from the Italian Ministry of University FIRB, Compagnia di San Paolo to MP, the Ministry of Health of Italy ‘Ricerca Corrente’ and ‘Ricerca Finalizzata’ to MP and GMF, AIRC to MP and MC, and Telethon to GMF. The support of the EU grant ‘Transpath ‘ Marie Curie project to MP is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Fimia, G., Corazzari, M., Antonioli, M. et al. Ambra1 at the crossroad between autophagy and cell death. Oncogene 32, 3311–3318 (2013). https://doi.org/10.1038/onc.2012.455
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.455
Keywords
This article is cited by
-
MYC-dependent MiR-7-5p regulated apoptosis and autophagy in diffuse large B cell lymphoma by targeting AMBRA1
Molecular and Cellular Biochemistry (2024)
-
High-dose-androgen-induced autophagic cell death to suppress the Enzalutamide-resistant prostate cancer growth via altering the circRNA-BCL2/miRNA-198/AMBRA1 signaling
Cell Death Discovery (2022)
-
Ambra1 regulates apoptosis and chemosensitivity in breast cancer cells through the Akt-FoxO1-Bim pathway
Apoptosis (2022)
-
Ambra1 in cancer: implications for clinical oncology
Apoptosis (2022)
-
Nuclear factor I-C disrupts cellular homeostasis between autophagy and apoptosis via miR-200b-Ambra1 in neural tube defects
Cell Death & Disease (2021)