Abstract
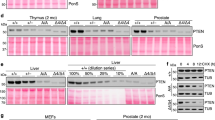
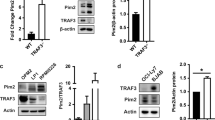
Pim kinases are found to be highly expressed in leukemia, lymphoma, prostate and pancreatic cancer. Bitransgenic mice overexpressing either Pim-1 or Pim-2 and c-Myc succumb to pre-B-cell lymphoma at a strikingly accelerated speed. Despite that Pim-1/Pim-2 has long been recognized as a strong synergistic partner with c-Myc in tumorigenesis, the mechanism underlying the synergism is still not well understood. Overexpression of Pim-1/Pim-2 kinase dramatically stabilizes c-Myc in vivo, and the stabilization is partially mediated by phosphorylation of c-Myc by Pim kinase on a novel site, Ser329. We provide evidence that Pim-2 is more efficient in directly phosphorylating c-Myc Ser329 to stabilize c-Myc. In contrast, we find that Pim-1 is more effective in mediating a decrease in c-Myc Thr58 phosphorylation and an increase in c-Myc Ser62 phosphorylation than in phosphorylating Ser329. In either case, through stabilizing c-Myc, Pim-1/Pim-2 kinases enhance the transcriptional activity of c-Myc. Also knocking down either Pim-1 or Pim-2 dramatically decreases the endogenous levels of c-Myc and thus, its transcriptional activity. Finally, coexpression of the Pim kinases and c-Myc enhances the transforming activity of c-Myc as does the phosphomimic mutant of c-Myc on Ser329. We conclude that these findings appear to explain at least in part the mechanism underlying the synergism between the Pim kinases and c-Myc in tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Allen JD, Verhoeven E, Domen J, van d V, Berns A . (1997). Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene 15: 1133–1141.
Amati B, Alevizopoulos K, Vlach J . (1998). Myc and the cell cycle. Front Biosci 3: d250–d268.
Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A . (1989). The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci USA 86: 8857–8861.
Breuer M, Wientjens E, Verbeek S, Slebos R, Berns A . (1991). Carcinogen-induced lymphomagenesis in pim-1 transgenic mice: dose dependence and involvement of myc and ras. Cancer Res 51: 958–963.
Bubman D, Guasparri I, Cesarman E . (2007). Deregulation of c-Myc in primary effusion lymphoma by Kaposi's sarcoma herpesvirus latency-associated nuclear antigen. Oncogene 26: 4979–4986.
Chen WW, Chan DC, Donald C, Lilly MB, Kraft AS . (2005). Pim family kinases enhance tumor growth of prostate cancer cells. Mol Cancer Res 3: 443–451.
Cowling VH, Cole MD . (2006). Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol 16: 242–252.
Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W . (1984). Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell 37: 141–150.
Dang CV, O'donnell KA, Juopperi T . (2005). The great MYC escape in tumorigenesis. Cancer Cell 8: 177–178.
De Both NJ, van der Feltz MJ, Mooren A, Vermaas D, Klaassen P, Rhijnsburger EH et al. (1989). Oncogene expression in Rauscher murine leukemia virus induced erythroid, myeloid and lymphoid cell lines. Leuk Res 13: 53–64.
Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K et al. (2001). Delineation of prognostic biomarkers in prostate cancer. Nature 412: 822–826.
Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R et al. (2003). Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4: 223–238.
Friedmann M, Nissen MS, Hoover DS, Reeves R, Magnuson NS . (1992). Characterization of the protooncogene Pim-1-kinase-activity and substrate recognition sequence. Arch Biochem Biophys 298: 594–601.
Hoover D, Friedmann M, Reeves R, Magnuson NS . (1991). Recombinant human Pim-1 protein exhibits serine threonine kinase-activity. J Biol Chem 266: 14018–14023.
Hurlin PJ, Huang J . (2006). The MAX-interacting transcription factor network. Semin Cancer Biol 16: 265–274.
Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N . (2006). Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res 66: 6741–6747.
Liu J, Martin HJ, Liao G, Hayward SD . (2007). The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J Virol 81: 10451–10459.
Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JSC . (2006). Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-X-L. BMC Cell Biol 7: 1.
Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G et al. (2006). Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 2: 1572–1581.
Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J et al. (2004). Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol 24: 6104–6115.
Moroy T, Verbeek S, Ma A, Achacoso P, Berns A, Alt F . (1991). E mu N- and E mu L-myc cooperate with E mu pim-1 to generate lymphoid tumors at high frequency in double-transgenic mice. Oncogene 6: 1941–1948.
Pelengaris S, Khan M, Evan G . (2002). c-MYC: more than just a matter of life and death. Nat Rev Cancer 2: 764–776.
Peng C, Knebel A, Morrice NA, Li X, Barringer K, Li J et al. (2007). Pim kinase substrate identification and specificity. J Biochem (Tokyo) 141: 353–362.
Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR . (2000). Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514.
Selten G, Cuypers HT, Berns A . (1985). Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J 4: 1793–1798.
van der Lugt NM, Domen J, Verhoeven E, Linders K, van der GH, Allen J et al. (1995). Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J 14: 2536–2544.
van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T et al. (1989). Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56: 673–682.
van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der GH, Berns A . (1991). Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65: 737–752.
Vita M, Henriksson M . (2006). The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16: 318–330.
Wanzel M, Herold S, Eilers M . (2003). Transcriptional repression by Myc. Trends Cell Biol 13: 146–150.
Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G et al. (2004). A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 6: 308–318.
Zhang Y, Wang Z, Magnuson NS . (2007). Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res 5: 909–922.
Zippo A, De Robertis A, Serafini R, Oliviero S . (2007). PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Bio 9: 932–944.
Acknowledgements
This study was supported by grant NIH R01-CA104470. We thank Dr Mark I Greene (School of Medicine, University of Pennsylvania) for providing us the luciferase reporter plasmid, pM4 min-luciferase.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Zhang, Y., Wang, Z., Li, X. et al. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 27, 4809–4819 (2008). https://doi.org/10.1038/onc.2008.123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2008.123
Keywords
This article is cited by
-
A promising natural product in diffuse large B-cell lymphoma therapy by targeting PIM1
Annals of Hematology (2024)
-
CDK9 inhibition induces epigenetic reprogramming revealing strategies to circumvent resistance in lymphoma
Molecular Cancer (2023)
-
Targeting Pim kinases in hematological cancers: molecular and clinical review
Molecular Cancer (2023)
-
Novel dual-targeting c-Myc inhibitor D347-2761 represses myeloma growth via blocking c-Myc/Max heterodimerization and disturbing its stability
Cell Communication and Signaling (2022)
-
Receptor-interacting protein kinase 2 (RIPK2) stabilizes c-Myc and is a therapeutic target in prostate cancer metastasis
Nature Communications (2022)