Abstract
Broadly neutralizing antibodies (bNAbs) against HIV-1 Env V1V2 arise in multiple donors. However, atomic-level interactions had previously been determined only with antibodies from a single donor, thus making commonalities in recognition uncertain. Here we report the cocrystal structure of V1V2 with antibody CH03 from a second donor and model Env interactions of antibody CAP256-VRC26 from a third donor. These V1V2-directed bNAbs used strand-strand interactions between a protruding antibody loop and a V1V2 strand but differed in their N-glycan recognition. Ontogeny analysis indicated that protruding loops develop early, and glycan interactions mature over time. Altogether, the multidonor information suggested that V1V2-directed bNAbs form an 'extended class', for which we engineered ontogeny-specific antigens: Env trimers with chimeric V1V2s that interacted with inferred ancestor and intermediate antibodies. The ontogeny-based design of vaccine antigens described here may provide a general means for eliciting antibodies of a desired class.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Sequence Read Archive
Referenced accessions
NCBI Reference Sequence
Protein Data Bank
Sequence Read Archive
References
Hraber, P. et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014).
Burton, D.R. et al. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5, 233–236 (2004).
Haynes, B.F., Kelsoe, G., Harrison, S.C. & Kepler, T.B. B-cell–lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30, 423–433 (2012).
Kwong, P.D. & Mascola, J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012).
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010).
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010).
Scheid, J.F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Zhou, T. et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258 (2013).
Zhou, T. et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 Supersite in 14 donors. Cell 161, 1280–1292 (2015).
Dosenovic, P. et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015).
Jardine, J.G. et al. HIV-1 vaccines: priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015).
Kong, L. et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20, 796–803 (2013).
Bonsignori, M. et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85, 9998–10009 (2011).
Doria-Rose, N.A. et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014).
Walker, L.M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009).
Walker, L.M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Sok, D. et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. USA 111, 17624–17629 (2014).
Pancera, M. et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84, 8098–8110 (2010).
Pejchal, R. et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. USA 107, 11483–11488 (2010).
McLellan, J.S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011).
Doria-Rose, N.A. et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. 86, 8319–8323 (2012).
Julien, J.P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. USA 110, 4351–4356 (2013).
Pancera, M. et al. N332-directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One 8, e55701 (2013).
Pancera, M. et al. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat. Struct. Mol. Biol. 20, 804–813 (2013).
Sanders, R.W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Ross, S.A., Sarisky, C.A., Su, A. & Mayo, S.L. Designed protein G core variants fold to native-like structures: sequence selection by ORBIT tolerates variation in backbone specification. Protein Sci. 10, 450–454 (2001).
Fazi, B. et al. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis. J. Biol. Chem. 277, 5290–5298 (2002).
McLellan, J.S. et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340, 1113–1117 (2013).
Zhou, T. et al. Transplanting supersites of HIV-1 vulnerability. PLoS One 9, e99881 (2014).
Do Kwon, Y. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Badger, J. et al. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60, 787–796 (2005).
Tyndall, J.D. et al. Macrophage migration inhibitory factor covalently complexed with phenethyl isothiocyanate. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 999–1002 (2012).
Pancera, M. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014).
Pan, R., Gorny, M.K., Zolla-Pazner, S. & Kong, X.P. The V1V2 region of HIV-1 gp120 forms a five-stranded beta barrel. J. Virol. 89, 8003–8010 (2015).
Briney, B.S., Willis, J.R. & Crowe, J.E. Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One 7, e36750 (2012).
Briney, B.S., Willis, J.R., Hicar, M.D., Thomas, J.W. II. & Crowe, J.E. Jr. Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology 137, 56–64 (2012).
Zhu, J. et al. Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. Proc. Natl. Acad. Sci. USA 110, 6470–6475 (2013).
Shi, B. et al. Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST. Theor. Biol. Med. Model. 11, 30 (2014).
Liao, H.X. et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38, 176–186 (2013).
Munro, J.B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014).
Sanders, R.W. et al. HIV-1 vaccines: HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223 (2015).
Meffre, E. et al. Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J. Clin. Invest. 108, 879–886 (2001).
Bhiman, J.N. et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 21, 1332–1336 (2015).
West, A.P. Jr., Diskin, R., Nussenzweig, M.C. & Bjorkman, P.J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. USA 109, E2083–E2090 (2012).
McGuire, A.T. et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210, 655–663 (2013).
Majeed, S. et al. Enhancing protein crystallization through precipitant synergy. Structure 11, 1061–1070 (2003).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007).
Calarese, D.A. et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003).
Doria-Rose, N.A. et al. A new member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. doi: 10.1128/JVI.01791-15 (14 October 2015).
Huang, J. et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature 515, 138–142 (2014).
Scheid, J.F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009).
Chen, L. et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326, 1123–1127 (2009).
Kwong, P.D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Killikelly, A. et al. Thermodynamic signatures of the antigen binding site of mAb 447-52D targeting the third variable region of HIV-1 gp120. Biochemistry 52, 6249–6257 (2013).
Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012).
Montefiori, D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485, 395–405 (2009).
Shu, Y. et al. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25, 1398–1408 (2007).
Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Guttman, M. et al. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22, 974–984 (2014).
Xiang, Z. & Honig, B. Extending the accuracy limits of prediction for side-chain conformations. J. Mol. Biol. 311, 421–430 (2001).
Petrey, D. et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53 (suppl. 6), 430–435 (2003).
Ye, J., Ma, N., Madden, T.L. & Ostell, J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013).
Sievers, F. & Higgins, D.G. Clustal omega. Curr. Protoc. Bioinformatics 48, 3.13.1–3.13.16 (2014).
Xiang, Z., Soto, C.S. & Honig, B. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. USA 99, 7432–7437 (2002).
Wriggers, W., Milligan, R.A. & McCammon, J.A. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 (1999).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Acknowledgements
We thank members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, NIH, for discussions and comments on the manuscript, and Weill Cornell Medical College, The Scripps Research Institute, Academic Medical Center and the HIV Vaccine Research and Design team comprising investigators from these three institutions for their contributions to the design and validation of near-native mimicry for soluble BG505 SOSIP.664 trimers. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson and L. Zhang for contributions to HIV-1-Env plasmids used in neutralization assessments. We thank R. Sanders for providing the PGDM1400–1412 sequences and the International AIDS Vaccine Initiative (IAVI) for PG9, PG16 and PGT141–145. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, US National Institute of Allergy and Infectious Diseases (NIAID) (to A.B.M., J.R. Mascola and P.D.K.); the Division of AIDS, NIAID, NIH (1U01-AI116086-01 to P.L.M., L.M., J.R. Mascola and P.D.K.; R21-AI112389 to K.K.L.); the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (OPP1033102 to K.K.L.); IAVI; and the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery grant (CHAVI-ID; UM1 AI100645 to M.B. and B.F.H.). This project was funded in part with Federal funds to U.B. from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38. Modeling and molecular dynamics were carried out through the NIH's Biowulf computing cluster.
Author information
Authors and Affiliations
Contributions
J.G. headed the determination of the V1V2-bound CH03 and CH04 crystal structures, revertant neutralization strategy and chimeric SOSIP design and assessment, and assisted with VRC26 model development; C.S. headed the next-generation-sequencing lineage analysis and VRC26 modeling; M.M.Y. assisted with crystallization. T.M.D., M.G. and K.K.L. performed HDX experiments and analyzed the data; R.T.B. and M.K.L. and J.R. Mascola assessed neutralization breadth; S.N., M.C. and A.B.M. performed antigenic analyses; G.-Y.C. performed frequentist probability analysis; B.J.D., J.R. McDaniel, G.G., X.W., J.C.M. and J.R. Mascola and NISC contributed next-generation sequencing data; J.G., C.S., M.P., N.A.D.-R., M.J.E., M.C.J. and B.Z. contributed to paratope mapping; A.D. expressed V1V2 scaffold and antibodies; J.G., I.S.G., Y.Y. and S.L. contributed to V1V2 scaffold design and assessment; C.S., T.M.L. and J.G. contributed to MDFF analysis; M.P. assisted with chimeric SOSIP design; J.S. assisted with figure conception and design; U.B. performed EM; T.Z. and M.G.J. contributed to reverted VRC01 experiments; M.B. and B.F.H. contributed CH0219 materials; P.L.M. and L.M. contributed CAP256 materials; J.G., C.S., L.S. and P.D.K. assembled and wrote the paper, on which all principal investigators commented.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
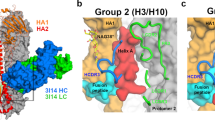
Supplementary Figure 1 Design and crystallization of trimeric scaffolded V1V2 with V1V2-directed broadly neutralizing antibodies.
(a) Using the trimeric orientation of V1V2 on the viral spike we used structure-based design to stabilize the domains on trimeric scaffolds. (b) Constructs were expressed in GnTI- cells by transient transfection in 96 well plates and screening was carried out using the trimer-specific antibodies PGT145 and VRC26. PGT145 binding was observed for 4 wells consisting of 2 different scaffolds, PDB IDs 1VH8 (yellow) and 4F2K (green). (c) Design examples of scaffolds which were bound by PGT145, 4F2K with various linker lengths at the c-terminal and 1VH8 which incorporated the V1V2 domain internally on the molecule. (d) These two and several others which showed good binding to PG9 were expressed at the 1 liter level and tested for binding by ELISA or SPR (fit in red). (e) The promising 1VH8 scaffold was optimized further by screening through 12 different HIV-1 strains and crystals were obtained for CH03 and CH04 bound to 1VH8CAP256SU and 1VH8A244 respectively.
Supplementary Figure 2 CH04 structure with PDB1VH8–scaffolded V1V2 from HIV-1 strain A244.
(a) The asymmetric unit of the crystal contains two fabs and two scaffolds. The structure was solved to 4.2 Å in space group P63, although significant twinning was observed. (b) The symmetry mates of one the molecule are shown as represented in figure 1 showing 3 fabs bound to the trimer in the biological assembly.
Supplementary Figure 3 Modeling the CAP256-VRC26 structure with V1V2.
(a) A model of VRC26.09 obtained through a combination of molecular dynamics (MD) and molecular dynamics flexible-fitting (MDFF) is shown with a negative stain 3D reconstruction of the fab complexed with BG505 SOSIP (EMD #5856). (b) HDX-MS is shown for BG505 SOSIP unliganded or in complex with VRC26.03. The difference between the bound and unbound data are plotted on one lobe of the trimer with regions of increased (blue) and decreased protection (red) mapped onto the structure (right panel) highlighting the region that VRC26.03 binds is similar to that seen in PG9, PG16, CH03 and CH04 complex crystal structures. (c) Paratope mapping of CAP256-VRC26 by arginine scanning. The left panel highlights the entire region scanned in green. Below, centroids of the introduced arginines are shown as red spheres. (right) Arginine variants were screened for neutralization across a panel of nine strains from multiple clades. The difference in neutralization compared to the WT antibody are shown in the table with differences greater than 20 fold indicated in red. Variants that significantly knocked down or knocked out neutralization were primarily localized to the CDR H3.
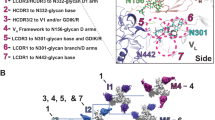
Supplementary Figure 4 Sequence features and structural role of the CDR H3 DDY motif.
(a) Sequence features of V1V2 bNAbs. Remarkably, although they use different germ-line genes, the amino acid sequence of the V-gene from donors CAP256 and IAVI24 are virtually identical. (b) Alignment of the CDR H3s from PG9 and CAP256-VRC26.09. The YYD motif is highlighted in red. (c) Structures of PG9 and VRC26.09 are aligned in the upper panel. The sulfated tyrosines of PG9 make electrostatic contacts with positively charged residues on the C-strand of the protomer with which the antibody is making main chain contacts. VRC26.09, however makes contacts with the positively charged side chains of each of the neighboring protomers at residue 169. Note that VRC26.09 also contains electrostatic interactions with the positively charged residues on the C strand it makes main chain contacts with (including residue 169), however it does not employ the YYD motif for this (Figure 5). We note that the VRC26.09 structure is a model and therefore the exact contacts may differ, for instance residue Arg166 of the neighboring V1V2 protomer is within range to make contacts with the sulfated tyrosine by selecting an alternative rotamer.
Supplementary Figure 5 NGS trees for donors IAVI 84 and CH0219.
(a) Sequence identity to neutralizing antibody PGT145 versus germline divergence to the germline gene IgHV1-8 for the Intra-donor positive data set. Sequence identity and germline divergence are expressed as a percentage. (b) Maximum likelihood tree for donor IAVI84 is shown colored according to the PGDM1400 series (blue) or the PGT141 series (green). (c-d) NGS tree for lineage CH01-CH04 from donor CH0219. (c) Sequence identity to neutralizing antibody CH04 versus germline divergence to the germline gene IgHV3-20 for the Illumina NGS data set. Sequence identity and germline divergence are expressed as a percentage. The corresponding sequence identity and germline divergence for the CH04 antibody is shown on the figure (note: there are no sequences in the NGS data sets with 100% identity to CH04). (d) Histogram distribution of CDR H3 lengths by V-germline (upper panel) and by V and J-germlines (lower panel). (e) Maximum likelihood tree for donor CH0219 is shown colored according to the method used to obtain the sequences with 454 in green and Illumina in blue.
Supplementary Figure 6 Somatic hypermutation and glycan recognition of V1V2-directed antibodies in donors CH0219 and IAVI24.
(a) V1V2 glycan interactions with PG16 and CH03 are shown. All residues on the antibodies which interact through hydrogen binding or have greater than 10% of their surface area buried by a glycan are shown as surface representation. Residues which are mutated from germline are colored red. Note that the atomic interactions with the glycan of the neighboring protomer for PG16 are not known. (b) Germline and mature sequences are shown for donors IAVI24 and CH0219. Residues that interact with glycan in the mature antibody structures (displayed as surface representation in (a)) are highlighted in grey. Somatically matured residues which interact with glycans are shown in red. 12 out of 21 residues that interact with glycans were not present in the earliest calculated intermediate for PG16 (2 positions are unknown and counted in the 12). 7 out of 24 residues that interact with glycans were not present in the earliest calculated intermediate for CH03.
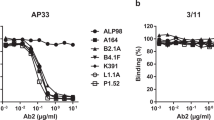
Supplementary Figure 7 Antigenic characterization of BG505 DS-SOSIP.664 chimeras with V1V2 regions from diverse HIV-1 strains.
(a) Strains of HIV-1 colored according to the representative dots in Figure 6. Sequences of the V1V2 domain (residue 126-196) from each strain are shown on the right with secondary structure derived from the BG505 structure shown below (b) MSD-ECLIA shows that the V1V2 chimeric BG505.SOSIPs behave antigenically similar to the native strain. Very low V3-directed antibody binding is seen following negative selection, comparable to levels of the BG505.SOSIP control. The constructs used here contain a D368R mutation to knock out CD4 binding which also knocks out VRC01 binding in this assay. Notably the trimers display variable levels of binding to PGT145 and VRC26, anticipated from their neutralization profiles of each strain. (c) ELISA experiments run in triplicate show specific strains bind reverted V1V2-directed antibodies better than others, even when mature binding is similar. With few exceptions, the binding correlates with the neutralization potency of each antibody. Strains are colored as in Fig. 6.
Supplementary Figure 8 Neutralization by germline-reverted VRC01-class antibodies identifies strains capable of interacting with early-lineage members.
(a) Strains neutralized by multiple germline-reverted VRC01-class antibodies were highlighted with colors. (b) Neutralization titers, loop D and loop V5 sequences and their potential glycosylation sites for strains neutralized by VRC01 germline antibodies were listed. (c) Location of loop D, V5 and CD4-binding loop on the HIV-1 Env.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 2042 kb)
Rights and permissions
About this article
Cite this article
Gorman, J., Soto, C., Yang, M. et al. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol 23, 81–90 (2016). https://doi.org/10.1038/nsmb.3144
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3144
This article is cited by
-
Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies
Nature Reviews Immunology (2023)
-
Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization
Nature Structural & Molecular Biology (2023)
-
Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site
Nature Communications (2023)
-
Differential V2-directed antibody responses in non-human primates infected with SHIVs or immunized with diverse HIV vaccines
Nature Communications (2022)
-
Human antibodies to SARS-CoV-2 with a recurring YYDRxG motif retain binding and neutralization to variants of concern including Omicron
Communications Biology (2022)