Abstract
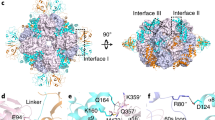
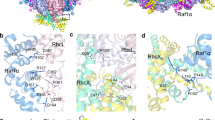
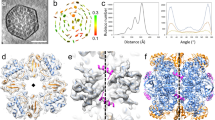
The form I Rubisco of autotrophic bacteria, algae and plants is a complex of eight large (RbcL) and eight small (RbcS) subunits. It fixes atmospheric CO2 in the dark reaction of photosynthesis. As shown for the cyanobacterial enzyme, folding of the RbcL subunits is mediated by the GroEL–GroES chaperonin system, and assembly requires the specialized chaperone RbcX, a homodimer of ~15-kDa subunits. Here we present the 3.2-Å crystal structure of a Rubisco assembly intermediate, consisting of the RbcL8 core with eight RbcX2 molecules bound. The structure reveals the molecular mechanism by which RbcX2 mediates oligomeric assembly. Specifically, RbcX2 provides positional information for proper formation of antiparallel RbcL dimers, thereby preventing RbcL–RbcL misalignment and off-pathway aggregation. The RbcL8(RbcX2)8 structure also suggests that RbcS functions by stabilizing the '60s loop' of RbcL in the catalytically active conformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Field, C.B. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998).
Andersson, I. Catalysis and regulation in Rubisco. J. Exp. Bot. 59, 1555–1568 (2008).
Tabita, F.R. et al. Function, structure, and evolution of the Rubisco-like proteins and their Rubisco homologs. Microbiol. Mol. Biol. Rev. 71, 576–599 (2007).
Tabita, F.R. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60, 1–28 (1999).
Andersson, I. & Taylor, T.C. Structural framework for catalysis and regulation in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414, 130–140 (2003).
Newman, J., Branden, C.I. & Jones, T.A. Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301. Acta Crystallogr. D Biol. Crystallogr. 49, 548–560 (1993).
Andrews, T.J. & Lorimer, G.H. Rubisco: structure, mechanisms, and prospects for improvement. in The Biochemistry of Plants: A Comprehensive Treatise, 131–218 (Academic Press, San Diego, California, USA, 1987).
Gutteridge, S. & Gatenby, A.A. Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell 7, 809–819 (1995).
Spreitzer, R.J. & Salvucci, M.E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 (2002).
Andrews, T.J. & Whitney, S.M. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 414, 159–169 (2003).
Spreitzer, R.J., Peddi, S.R. & Satagopan, S. Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl. Acad. Sci. USA 102, 17225–17230 (2005).
Whitney, S.M., Houtz, R.L. & Alonso, H. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35 (2011).
Goloubinoff, P., Christeller, J.T., Gatenby, A.A. & Lorimer, G.H. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and MgATP. Nature 342, 884–889 (1989).
Brinker, A. et al. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233 (2001).
Hartl, F.U. & Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 (2009).
Saschenbrecker, S. et al. Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129, 1189–1200 (2007).
Liu, C. et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463, 197–202 (2010).
Tanaka, S., Sawaya, M.R., Kerfeld, C.A. & Yeates, T.O. Structure of the Rubisco chaperone RbcX from Synechocystis sp PCC6803. Acta Crystallogr. D Biol. Crystallogr. 63, 1109–1112 (2007).
Tarnawski, M., Gubernator, B., Kolesinski, P. & Szczepaniak, A. Heterologous expression and initial characterization of recombinant RbcX protein from Thermosynechococcus elongatus BP-1 and the role of RbcX in Rubisco assembly. Acta Biochim. Pol. 55, 777–785 (2008).
Curmi, P.M., Cascio, D., Sweet, R.M., Eisenberg, D. & Schreuder, H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-Å resolution. J. Biol. Chem. 267, 16980–16989 (1992).
Mueller-Cajar, O. & Whitney, S.M. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 414, 205–214 (2008).
Spreitzer, R.J. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414, 141–149 (2003).
Duff, A.P., Andrews, T.J. & Curmi, P.M. The transition between the open and closed states of Rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 298, 903–916 (2000).
Read, B.A. & Tabita, F.R. Amino acid substitutions in the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase that influence catalytic activity of the holoenzyme. Biochemistry 31, 519–525 (1992).
Flachmann, R., Zhu, G., Jensen, R.G. & Bohnert, H.J. Mutations in the small subunit of ribulose-1,5-bisphosphate carboxylase/ oxygenase increase the formation of the misfire product xylulose-1,5-bisphosphate. Plant Physiol. 114, 131–136 (1997).
Genkov, T. & Spreitzer, R.J. Highly conserved small subunit residues influence Rubisco large subunit catalysis. J. Biol. Chem. 284, 30105–30112 (2009).
Nishihara, K., Kanemori, M., Yanagi, H. & Yura, T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 66, 884–889 (2000).
Park, E.S., Fenton, W.A. & Horwich, A.L. Disulfide formation as a probe of folding in GroEL-GroES reveals correct formation of long-range bonds and editing of incorrect short-range ones. Proc. Natl. Acad. Sci. USA 104, 2145–2150 (2007).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. Scala. Joint CCP4 + ESF-EACBM Newsletter on Prot. Crystallogr. 33, 22–24 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
French, G. & Wilson, K. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978).
Vagin, A.A. & Isupov, M.N. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 57, 1451–1456 (2001).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
We thank S. Saschenbrecker for performing initial crystallization trials and the Joint Structural Biology Group staff at the European Molecular Biology Laboratory and European Synchrotron Radiation Facility, Grenoble, France, for their support at beamlines ID29, ID23-2 and ID23-1.
Author information
Authors and Affiliations
Contributions
A.B. crystallized the Rubisco assembly intermediate, solved the structure and designed the mutants. A.S.-W. executed the biochemical study. A.B., A.S.-W., F.U.H. and M.H.-H. contributed to experimental design, analysis and interpretation of data. A.B., F.U.H. and M.H.-H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Table 1 (PDF 417 kb)
Rights and permissions
About this article
Cite this article
Bracher, A., Starling-Windhof, A., Hartl, F. et al. Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco. Nat Struct Mol Biol 18, 875–880 (2011). https://doi.org/10.1038/nsmb.2090
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2090
This article is cited by
-
Scaffolding protein CcmM directs multiprotein phase separation in β-carboxysome biogenesis
Nature Structural & Molecular Biology (2021)
-
Is RAF1 protein from Synechocystis sp. PCC 6803 really needed in the cyanobacterial Rubisco assembly process?
Photosynthesis Research (2017)
-
From cyanochemicals to cyanofactories: a review and perspective
Microbial Cell Factories (2016)
-
Opposing effects of folding and assembly chaperones on evolvability of Rubisco
Nature Chemical Biology (2015)
-
Structure and mechanism of the Rubisco-assembly chaperone Raf1
Nature Structural & Molecular Biology (2015)