Abstract
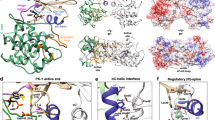
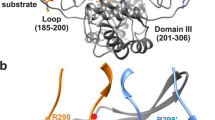
Biochemical studies indicate that dimerization is required for the catalytic activity of herpesvirus proteases, whereas structural studies show a complete active site in each monomer, away from the dimer interface. Here we report kinetic, biophysical and crystallographic characterizations of structure-based mutants in the dimer interface of human cytomegalovirus (HCMV) protease. Such mutations can produce a 1,700-fold reduction in the kcat while having minimal effects on the Km. Dimer stability is not affected by these mutations, suggesting that dimerization itself is insufficient for activity. There are large changes in monomer conformation and dimer organization of the apo S225Y mutant enzyme. However, binding of an activated peptidomimetic inhibitor induced a conformation remarkably similar to the wild type protease. Our studies suggest that appropriate dimer formation may be required to indirectly stabilize the protease oxyanion hole, revealing a novel mechanism for dimerization to regulate enzyme activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fields, B.N., Knipe, D.M. & Howley, P.M. Fields virology (Lippincott-Raven Press, New York; 1996).
Gibson, W., Welch, A.R. & Hall, M.R.T. Assemblin, a herpes virus serine maturational proteinase and new molecular target for antivirals. Persp. Drug Discovery 2, 413–426 (1995).
Waxman, L. & Darke, P.L. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir. Chem. Chemother. 11, 1–22 (2000).
Tong, L. et al. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383, 272–275 (1996).
Qiu, X. et al. Unique fold and active site in cytomegalovirus protease. Nature 383, 275–279 (1996).
Shieh, H.-S. et al. Three-dimensional structure of human cytomegalovirus protease. Nature 383, 279–282 (1996).
Chen, P. et al. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86, 835–843 (1996).
Qiu, X. et al. Crystal structure of varicella-zoster virus protease. Proc. Natl. Acad. Sci. USA 94, 2874–2879 (1997).
Hoog, S.S. et al. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry 36, 14023–14029 (1997).
Reiling, K.K., Pray, T.R., Craik, C.S. & Stroud, R.M. Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements. Biochemistry 39, 12796–12803 (2000).
Tong, L. et al. Conserved mode of peptidomimetic inhibition and substrate recognition of human cytomegalovirus protease. Nature Struct. Biol. 5, 819–826 (1998).
Tong, L. et al. Experiences from the structure determination of human cytomegalovirus protease. Acta Crystallogr. D 53, 682–690 (1997).
Pinko, C. et al. Single-chain recombinant human cytomegalovirus protease. J. Biol. Chem. 270, 23634–23640 (1995).
Holskin, B.P. et al. A continuous fluorescence-based assay of human cytomegalovirus protease using a peptide substrate. Anal. Biochem. 227, 148–155 (1995).
Cole, J.L. Characterization of human cytomegalovirus protease dimerization by analytical centrifugation. Biochemistry 35, 15601–15610 (1996).
Darke, P.L. et al. Active human cytomegalovirus protease is a dimer. J. Biol. Chem. 271, 7445–7449 (1996).
Margosiak, S.A., Vanderpool, D.L., Sisson, W., Pinko, C. & Kan, C.-C. Dimerization of the human cytomegalovirus protease: kinetic and biochemical characterization of the catalytic homodimer. Biochemistry 35, 5300–5307 (1996).
Tong, L. Combined molecular replacement. Acta Crystallogr. A 52, 782–784 (1996).
Jogl, G., Tao, X., Xu, Y. & Tong, L. COMO: a program for combined molecular replacement. Acta Crystallogr. D 57, 1127–1134 (2001).
Liang, P.-H. et al. Site-directed mutagenesis probing the catalytic role of arginines 165 and 166 of human cytomegalovirus protease. Biochemistry 37, 5923–5929 (1998).
Bonneau, P.R. et al. Evidence of a conformational change in the human cytomegalovirus protease upon binding of peptidyl-activated carbonyl inhibitors. Biochemistry 36, 12644–12652 (1997).
Schechter, I. & Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 (1967).
Ogilvie, W. et al. Peptidomimetic inhibitors of the human cytomegalovirus protease. J. Med. Chem. 40, 4113–4135 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Carson, M. Ribbon models of macromolecules. J. Mol. Graphics 5, 103–106 (1987).
Nicholls, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Evans, S.V. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11, 134–138 (1993).
Acknowledgements
We thank R. Olson for help with analytical ultracentrifugation experiments; C. Parish for help with CD measurements; M. Bailey for the synthesis of BILC408; Y. Xu, Z. Yang, C. Ogata and J. Berendzen for help with data collection at the synchrotron radiation source; W.W. Cleland for helpful discussions and the National Institutes of Health (grant to L.T.) for financial support. R.K. is supported by the training program in molecular biophysics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Batra, R., Khayat, R. & Tong, L. Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nat Struct Mol Biol 8, 810–817 (2001). https://doi.org/10.1038/nsb0901-810
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0901-810
This article is cited by
-
Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes
Nature Communications (2024)
-
Soluble expression, purification, and characterization of active recombinant human tissue plasminogen activator by auto-induction in E. coli
BMC Biotechnology (2015)
-
Inhibition of a viral enzyme by a small-molecule dimer disruptor
Nature Chemical Biology (2009)
-
Induced structure of a helical switch as a mechanism to regulate enzymatic activity
Nature Structural & Molecular Biology (2005)