Abstract
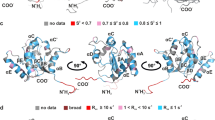
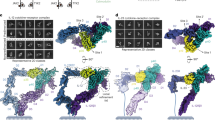
The structure of a folded core of IL-16 is similar to that of intracellular protein modules called PDZ domains. IL-16 is thus the first extracellular protein found to have a PDZ-like fold. However, it does not exhibit normal peptide binding properties of PDZ domains. This is due to alterations of the structure at the 'PDZ-like binding site' of IL-16 (the GLGF cleft): the GLGF cleft of IL-16 is much smaller than those of PDZ-domains and is additionally blocked with a tryptophan side chain at its center. Our experiments indicate also that IL-16 nonspecifically aggregates in solution; but formation of a homo-tetrameric protein is not required, in contrast to previous suggestions, for its chemo-attractant activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Center, D. M. & Cruikshank, W. W. Modulation of lymphocyte migration by human lymphokines (identification and characterization). J. Immunol. 128, 2563– 2568 (1982).
Cruikshank, W. W. & Center, D. M. Modulation of lymphocyte migration by human lymphokines (purification). J. Immunol. 128 , 2569– 2574 (1982).
Baier, M., Werner, A., Bannert, N., Metzner, K. & Kurth, R. HIV suppression by interleukin-16. Nature 378, – 563 (1995 ).
Rumsaeng, V., Cruikshank, W. W. & Foster, B. Human mast-cells produce the CD4(+) T-lymphocyte chemoattractant factor, IL-16 . J. Immunol. 159, 2904– 2910 (1997).
Cruikshank, W. W., Greenstein, J. L., Theodore, A. C. & Center, D. M. Lymphocyte chemoattractant factor induces CD4-dependent intracytoplasmic signaling in lymphocytes. J. Immunol. 146, 2928– 2934 (1991).
Center, D. M., Kornfeld, H. & Cruikshank, W. W. Interleukin 16 and its function as a CD4 ligand. Immunol. Today 17, 476– 481 (1996).
Baier, M. & Kurth, R. Fighting HIV-1 with IL-16. Nature Med. 3, 605– 606 ( 1997).
Bazan, J. F. & Schall, T. J. Interleukin-16 or not? Nature 381, 29– 30 ( 1996).
Mackewicz, C. E., Levy, J. A., Cruikshank, W. W., Kornfeld, H. & Center, D. M. Role of IL-16 in HIV replication . Nature 383, 488– 489 (1996).
Levy, J. A., Mackewicz, C. E. & Barker, E. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol. Today 17, 217– 224 (1996).
Zhou, P., Goldstein, S., Devadas, K., Tewari, D. & Notkins, A. L. Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: Inhibition of mRNA expression. Nature Med. 3, 659– 664 ( 1997).
Viglianti, G. A. et al. IL-16 anti-HIV-1 therapy. Nature Med. 3, 938 938 (1998).
Maciaszek, J. W. et al. IL-16 represses HIV-1 promoter activity. J. Immunol. 158, 5– 8 (1997).
Cruikshank, W. W. et al. Molecular and functional analysis of a lymphocyte chemoattractant factor: Association of biologic function with CD4 expression. Proc. Natl. Acad. Sci. USA 91, 5109– 5113 (1994).
Baier, M., Bannert, N., Werner, A., Lang, K. & Kurth, R. Molecular cloning, sequence, expression, and processing of the interleukin 16 precursor. Proc. Natl. Acad. Sci.USA 94, 5273– 5277 (1997).
Zhang, Y. et al. Processing and activation of pro-interleukin-16 by caspase-3. J. Biol. Chem. 273, 1144– 1149 (1998).
Farrow, N. A. et al. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984– 6003 ( 1994).
Edison, A. S., Abildgaard, F., Westler, W. M., Mooberry, E. S. & Markley, J. L. Practical introduction to theory and implementation of multinuclear, multidimensional nuclear magnetic resonance experiments. Meth. Enz. 239, 3– 79 ( 1994).
Zwahlen, C. et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage N-peptide/box B RNA complex. J. Am. Chem. Soc. 119, 6711– 6721 (1997).
Vriend, G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52– 56 (1990).
Ponting, C. P. & Phillips, C. DHR domains in syntrophins, neuronal NO synthases and other intracellular proteins Trends Biochem. Sci. 20, 102– 103 ( 1995).
Songyang, Z. et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73– 77 ( 1997).
Cabral, J. H., Petosa, C. & Sutcliffe, M. J. Crystal structure of a PDZ domain. Nature 382, 649– 652 (1996).
Shuker, S. B., Hajduk, P. J., Meadows, R. P. & Fesik, S. W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531– 1534 (1996).
Hu, J. S. & Bax, A. 01 angle information from a simple 2-dimensional NMR experiment that identifies trans 3JNC couplings in isotopically enriched proteins. J. Biomol. NMR 9, 323– 328 (1997).
Hu, J. S. & Bax, A. Determination of . angles and 01 angles in proteins from13C-13C three-bond J-couplings measured by 3-dimensional heteronuclear NMR. How planar is the peptide bond . J. Am. Chem. Soc. 119, 6360– 6368 (1997).
Vuister, G. W. et al. Measurement of homo- and heteronuclear J-couplings from quantitative J correlation . Meth. Enz. 239, 79– 105 (1994).
Brünger, A.T. X-PLOR, Version 3.1: System for X-ray Crystallography and NMR Yale Univ. Press, New Haven and London, (1992).
Wishart, D. S., Sykes, B. D., & Richards, F. M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311– 333 (1991).
Wüthrich, K. NMR of Proteins and Nucleic Acids (Wiley, New York, 1986).
Schultz, J. et al. Specific interactions between the syntrophin PDZ domain and voltage gated sodium channels. Nature Struct. Biol. 5, 19– 24 (1998).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51– 55 (1996).
Holak, T. A., Gondol, D., Otlewski, J. & Wilusz, T. Determination of the complete 3-dimensional structure of the trypsin-inhibitor from squash seeds in aqueous-solution by nuclear magnetic-resonance and a combination of distance geometry and dynamical simulated annealing. J. Mol. Biol. 210, 635– 648 ( 1989).
Doyle, D. A. et al. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85, 1067– 1076 (1996).
Acknowledgements
We thank R. Engh for stimulating discussions, F. Hesse for protein purification, H. Burtscher for cloning and expression, H. Popp for fermentation, M. Wozny for mass analysis and R. Rudolf for analytical ultracentrifugation. M. Z. is a recipient of the graduate scholarship from Verband der Chemischen Industrie.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mühlhahn, P., Zweckstetter, M., Georgescu, J. et al. Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site. Nat Struct Mol Biol 5, 682–686 (1998). https://doi.org/10.1038/1376
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/1376
This article is cited by
-
Pressuromodulation at the cell membrane as the basis for small molecule hormone and peptide regulation of cellular and nuclear function
Journal of Translational Medicine (2015)
-
Introduction of pro-interleukin-16 inhibits T-lymphoblastic leukemia growth in mice
Journal of Cancer Research and Clinical Oncology (2011)
-
Proteolytic cleavage of PDZD2 generates a secreted peptide containing two PDZ domains
EMBO reports (2003)