Key Points
-
Measuring the microbiome and/or microbiota is challenging; new bioinformatic techniques are becoming available that can help identify patterns in complex data sets
-
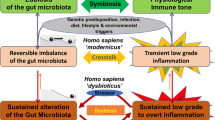
Numerous pre-clinical studies show that the microbiota has profound effects on the host immune system, which can influence the balance between tolerance and immunity
-
Microbial species and gene catalogues can help identify diagnostic biomarkers that correlate with outcomes in organ transplant recipients
-
Functional analyses of the microbiota will be required to characterize specific pathophysiological pathways and define therapeutic interventions
-
Clinical studies will be required to correlate features of the microbiota with the outcomes of organ transplantation, including graft survival, infection, rejection, recurrent disease, inflammation, alloreactivity and fibrosis
-
Challenges lie in translating associations between the microbiota and transplantation outcomes into definitive demonstrations of mechanism and causality, which are limitations that currently plague clinical investigation of the microbiota
Abstract
Each individual harbours a unique set of commensal microorganisms, collectively referred to as the microbiota. Notably, these microorganisms exceed the number of cells in the human body by 10-fold. This finding has accelerated a shift in our understanding of human physiology, with the realization that traits necessary for health are both encoded and influenced by the human genome and the microbiota. Our understanding of the aetiology of complex diseases has, therefore, evolved with increasing awareness that the human microbiota has an active and critical role in maintaining health and inducing disease. Indeed, findings from bioinformatic studies indicate that the microbiota and microbiome have multiple effects on the innate and adaptive immune systems, with effects on infection, autoimmune disease and cancer. In this Review, we first address the important statistical and informatics aspects that should be considered when characterizing the composition of microbiota. We next highlight the effects of the microbiota on the immune system and the implications of these effects on organ failure and transplantation. Finally, we reflect on the future perspectives for studies of the microbiota, including novel diagnostic tests and therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Savage, D. C. Microbial Ecology of the Gastrointestinal Tract. Annu. Rev. Microbiol. 31, 107–133 (1977).
National Research Council (US) Committee on Metagenomics: Challenges and Functional Applications. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. (National Academies Press [US], 2007).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Weng, M. & Walker, W. A. The role of gut microbiota in programming the immune phenotype. J. Dev. Orig. Health Dis. 4, 203–214 (2013).
Schwabe, R. F. & Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 13, 800–812 (2013).
Stecher, B. & Hardt, W.-D. The role of microbiota in infectious disease. Trends Microbiol. 16, 107–114 (2008).
Wu, H.-J. & Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 3, 4–14 (2012).
Fricke, W. F., Maddox, C., Song, Y. & Bromberg, J. S. Human microbiota characterization in the course of renal transplantation. Am. J. Transplant. 14, 416–427 (2014).
Oh, P. L. et al. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am. J. Transplant. 12, 753–762 (2012).
Jenq, R. R. et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 209, 903–911 (2012).
National Institutes of Health. Human microbiome project [online], (2015).
MetaHIT. Metagenomics of the human intestinal tract [online], (2012).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J. & Goodman, R. M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–249 (1998).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
RDP release 11. Sequence analysis tools [online], (2014).
Silva. High quality ribosomal RNA databases [online], (2014).
Green genes. 16S rDNA data and tools [online], (2013).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Schloss, P. D. & Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005).
Hill, T. C. J., Walsh, K. A., Harris, J. A. & Moffett, B. F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11 (2003).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Lozupone, C. A., Hamady, M., Kelley, S. T. & Knight, R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585 (2007).
Magurran, A. E. Measuring Biological Diversity (Wiley-Blackwell, 2004).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Romero, R. et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2, 4 (2014).
McGarigal, K., Landguth, E. & Stafford, S. Multivariate Statistics for Wildlife and Ecology Research (Springer, 2002).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Xia, L. C. et al. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst. Biol. 5 (Suppl. 2), S15 (2011).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Matsumoto, S., Setoyama, H. & Umesaki, Y. Differential induction of major histocompatibility complex molecules on mouse intestine by bacterial colonization. Gastroenterology 103, 1777–1782 (1992).
Lundin, A. et al. Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 10, 1093–1103 (2008).
Campos Canesso, M. et al. Comparing the effects of acute alcohol consumption in germ-free and conventional mice: the role of the gut microbiota. BMC Microbiol. 14, 240 (2014).
Seki, E. et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 (2007).
Turley, S. J., Lee, J. W., Dutton-Swain, N., Mathis, D. & Benoist, C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc. Natl Acad. Sci. USA 102, 17729–17733 (2005).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Hänninen, A. et al. Islet beta-cell-specific T cells can use different homing mechanisms to infiltrate and destroy pancreatic islets. Am. J. Pathol. 170, 240–250 (2007).
Molloy, M., Bouladoux, N. & Belkaid, Y. Intestinal Microbiota: Shaping local and systemic immune responses. Semin. Immunol. 24, 58–66 (2012).
Ridaura, V. K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Glaister, J. R. Factors affecting the lymphoid cells in the small intestinal epithelium of the mouse. Int. Arch. Allergy Immunol. 45, 719–730 (1973).
Pollard, M. & Sharon, N. Responses of the peyer's patches in germ-free mice to antigenic stimulation. Infect. Immun. 2, 96–100 (1970).
Hoshi, H. et al. Lymph follicles and germinal centers in popliteal lymph nodes and other lymphoid tissues of germ-free and conventional rats. Tohoku J. Exp. Med. 166, 297–307 (1992).
Clarke, T. B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Rivas, M. N. et al. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J. Clin. Invest. 122, 1933–1947 (2012).
Sakaguchi, S. et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 212, 8–27 (2006).
Chinen, T., Volchkov, P. Y., Chervonsky, A. V. & Rudensky, A. Y. A critical role for regulatory T cell-mediated control of inflammation in the absence of commensal microbiota. J. Exp. Med. 207, 2323–2330 (2010).
Bang, C., Weidenbach, K., Gutsmann, T., Heine, H. & Schmitz, R. A. The intestinal archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE 9, e99411 (2014).
Stepankova, R. et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211 (2007).
Niess, J. H., Leithäuser, F., Adler, G. & Reimann, J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J. Immunol. 180, 559–568 (2008).
Bermudez-Brito, M. et al. Human intestinal dendritic cells decrease cytokine release against salmonella infection in the presence of Lactobacillus paracasei upon TLR activation. PLoS ONE 7, e43197 (2012).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA http://dx.doi.org/10.1073/pnas.0804812105.
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Round, J. L. et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Dewhirst, F. E. et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65, 3287–3292 (1999).
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011).
Josefowicz, S. Z. et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482, 395–399 (2012).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Chehoud, C. et al. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc. Natl Acad. Sci. USA 110, 15061–15066 (2013).
Kurashima, Y. et al. The enzyme Cyp26b1 mediates inhibition of mast cell activation by fibroblasts to maintain skin-barrier homeostasis. Immunity 40, 530–541 (2014).
Nakamura, Y. et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 503, 397–401 (2013).
Canesso, M. C. C. et al. Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J. Immunol. 193, 5171–5180 (2014).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Shen, W. et al. Adaptive immunity to murine skin commensals. Proc. Natl Acad. Sci. USA 111, E2977–E2986 (2014).
Lai, Y. et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 (2009).
Fyhrquist, N. et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J. Allergy Clin. Immunol. 136, 1301–1309 (2014).
Volz, T. et al. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J. Invest. Dermatol. 134, 96–104 (2014).
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA 108, 5354–5359 (2011).
Gollwitzer, E. S. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647 (2014).
Nicolle, L. E. et al. Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin. Infect. Dis. 40, 643–654 (2005).
Xu, W. et al. Mini-review: perspective of the microbiome in the pathogenesis of urothelial carcinoma. Am. J. Clin. Exp. Urol. 2, 57–61 (2014).
Cheng, Y.-W. & Wong, S.-N. Diagnosing symptomatic urinary tract infections in infants by catheter urine culture. J. Paediatr. Child Health 41, 437–440 (2005).
Siddiqui, H., Nederbragt, A. J., Lagesen, K., Jeansson, S. L. & Jakobsen, K. S. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 11, 244 (2011).
Siddiqui, H., Lagesen, K., Nederbragt, A. J., Jeansson, S. L. & Jakobsen, K. S. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol. 12, 205 (2012).
Hilt, E. E. et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52, 871–876 (2014).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 (2008).
Endesfelder, D. et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes 63, 2006–2014 (2014).
Bauer, H., Horowitz, R. E., Levenson, S. M. & Popper, H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 42, 471–483 (1963).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Koren, O. et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl Acad. Sci. USA 108, 4592–4598 (2011).
Oh, J. Z. et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity 41, 478–492 (2014).
Andrade-Oliveira, V. et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J. Am. Soc. Nephrol. http://dx.doi.org/10.1681/ASN.2014030288.
Harkiolaki, M. et al. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity 30, 348–357 (2009).
Pantenburg, B., Heinzel, F., Das, L., Heeger, P. S. & Valujskikh, A. T cells primed by Leishmania major infection cross-react with alloantigens and alter the course of allograft rejection. J. Immunol. 169, 3686–3693 (2002).
Atkinson, M. A. et al. Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin-dependent diabetes. J. Clin. Invest. 94, 2125–2129 (1994).
McClain, M. T. et al. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat. Med. 11, 85–89 (2005).
Lunardi, C. et al. Systemic sclerosis immunoglobulin G autoantibodies bind the human cytomegalovirus late protein UL94 and induce apoptosis in human endothelial cells. Nat. Med. 6, 1183–1186 (2000).
Holdener, M. et al. Breaking tolerance to the natural human liver autoantigen cytochrome P450 2D6 by virus infection. J. Exp. Med. 205, 1409–1422 (2008).
Zhao, Z.-S., Granucci, F., Yeh, L., Schaffer, P. A. & Cantor, H. Molecular mimicry by herpes simplex virus-type 1: autoimmune disease after viral infection. Science 279, 1344–1347 (1998).
Bachmaier, K. et al. Chlamydia infections and heart disease linked through antigenic mimicry. Science 283, 1335–1339 (1999).
Westall, F. C. Molecular mimicry revisited: gut bacteria and multiple sclerosis. J. Clin. Microbiol. 44, 2099–2104 (2006).
Amedei, A. et al. Molecular mimicry between Helicobacter pylori antigens and H+, K+—adenosine triphosphatase in human gastric autoimmunity. J. Exp. Med. 198, 1147–1156 (2003).
Deshmukh, U. S. et al. HLA-DR3 restricted T cell epitope mimicry in induction of autoimmune response to lupus-associated antigen SmD. J. Autoimmun. 37, 254–262 (2011).
Kirvan, C. A., Swedo, S. E., Heuser, J. S. & Cunningham, M. W. Mimicry and autoantibody-mediated neuronal cell signaling in Sydenham chorea. Nat. Med. 9, 914–920 (2003).
Massa, M. et al. Self epitopes shared between human skeletal myosin and Streptococcus pyogenes M5 protein are targets of immune responses in active juvenile dermatomyositis. Arthritis Rheum. 46, 3015–3025 (2002).
Ciubotariu, R. et al. Persistent allopeptide reactivity and epitope spreading in chronic rejection of organ allografts. J. Clin. Invest. 101, 398–405 (1998).
Amir, A. L. et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood 115, 3146–3157 (2010).
Brehm, M. A. et al. Allografts stimulate cross-reactive virus-specific memory CD8 T cells with private specificity. Am. J. Transplant. 10, 1738–1748 (2010).
D'Orsogna, L. J. A. et al. Vaccine-induced allo-HLA-reactive memory T cells in a kidney transplantation candidate. Transplantation 91, 645–651 (2011).
Hudson, K. E., Lin, E., Hendrickson, J. E., Lukacher, A. E. & Zimring, J. C. Regulation of primary alloantibody response through antecedent exposure to a microbial T-cell epitope. Blood 115, 3989–3996 (2010).
Abt, M. C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Maier, S. et al. Inhibition of natural killer cells results in acceptance of cardiac allografts in CD28−/− mice. Nat. Med. 7, 557–562 (2001).
Morita, K. et al. Early chemokine cascades in murine cardiac grafts regulate T cell recruitment and progression of acute allograft rejection. J. Immunol. 167, 2979–2984 (2001).
Tanaka, K., Sawamura, S., Satoh, T., Kobayashi, K. & Noda, S. Role of the indigenous microbiota in maintaining the virus-specific CD8 memory T cells in the lung of mice infected with murine cytomegalovirus. J. Immunol. 178, 5209–5216 (2007).
Schwab, L. et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat. Med. 20, 648–654 (2014).
Taur, Y. et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182 (2014).
Hall, J. A. et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29, 637–649 (2008).
Hand, T. W. et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012).
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal Infection. Immunity 31, 772–786 (2009).
Vaziri, N. D. et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 83, 308–315 (2013).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Diaz, P. I. et al. Transplantation-associated long-term immunosuppression promotes oral colonization by potentially opportunistic pathogens without impacting other members of the salivary bacteriome. Clin. Vaccine Immunol. 20, 920–930 (2013).
Li, Q. R. et al. Reciprocal interaction between intestinal microbiota and mucosal lymphocyte in cynomolgus monkeys after alemtuzumab treatment. Am. J. Transplant. 13, 899–910 (2013).
Li, Q., Wang, C., Tang, C., He, Q. & Li, J. Lymphocyte depletion after alemtuzumab induction disrupts intestinal fungal microbiota in cynomolgus monkeys. Transplantation 98, 951–959 (2014).
Soave, R. Prophylaxis strategies for solid-organ transplantation. Clin. Infect. Dis. 33 (Suppl. 1), S26–S31 (2001).
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (2008).
Antunes, L. C. M. et al. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob. Agents Chemother. 55, 1494–1503 (2011).
Sekirov, I., Russell, S. L., Antunes, L. C. M. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Modi, S. R., Lee, H. H., Spina, C. S. & Collins, J. J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108, 4554–4561 (2011).
Bouatra, S. et al. The human urine metabolome. PLoS ONE 8, e73076 (2013).
Youngster, I. et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 312, 1772–1778 (2014).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2014).
Macdonald, W. A. et al. T cell allorecognition via molecular mimicry. Immunity 31, 897–908 (2009).
Author information
Authors and Affiliations
Contributions
All authors researched the data for the article, provided substantial contributions to discussions of its content, wrote the article and undertook review and/or editing of the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Bromberg, J., Fricke, W., Brinkman, C. et al. Microbiota—implications for immunity and transplantation. Nat Rev Nephrol 11, 342–353 (2015). https://doi.org/10.1038/nrneph.2015.70
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneph.2015.70
This article is cited by
-
Role of metagenomics in prospecting novel endoglucanases, accentuating functional metagenomics approach in second-generation biofuel production: a review
Biomass Conversion and Biorefinery (2023)
-
Diet simplification selects for high gut microbial diversity and strong fermenting ability in high-altitude pikas
Applied Microbiology and Biotechnology (2018)
-
Gut microbiota and its implications in small bowel transplantation
Frontiers of Medicine (2018)