Key Points
-
The human gastrointestinal tract (GIT) is a complex ecosystem, the bacterial components (microbiota) of which are thought to have a significant role in normal gut function and in maintaining host health. The human gut microbiota include health-promoting indigenous species such as Bifidobacterium and Lactobacillus, also referred to as probiotic bacteria. Probiotic bacteria are commonly consumed live as dietary supplements.
-
The molecular mechanisms by which probiotic bacteria exert their health-promoting effects remain largely unclear. However, the advent of a novel scientific discipline, called probiogenomics, has recently provided new insights into the diversity and evolution of probiotic bacteria and has revealed the molecular basis of probiosis.
-
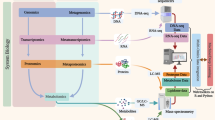
Probiogenomic efforts have shown how the genome content of bifidobacteria and lactobacilli reflect adaptations to the human intestinal niche. Genomic evidence for adaptations to the GIT includes metabolic features, such as the capacity for uptake of macromolecules and breakdown of undigested complex carbohydrates, and the ability to interact with the host through the production of cell-surface proteins that interact with the intestinal mucosa.
-
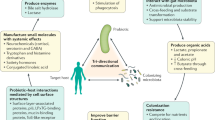
The interaction of probiotic bacteria with the host, as well as with other components of the human gut microbiota, is considered a key feature of probiosis. Bifidobacteria inducean expansion in the diversity of polysaccharides that are targeted for degradation by common intestinal bacteria (Bacteroides) and also inducethe expression of host genes that play a part in innate immunity.
-
Comparisons among completely sequenced bifidobacterial and lactobacilli genomes revealed that the main force that drives evolution in these genomes is horizontal gene transfer.
-
Probiotic bacteria are diverse and taxonomically heterogeneous groups of microorganisms, so the analysis of phyletic patterns — that is, patterns of gene presence or absence in a particular set of genomes — might be influenced by the evolutionary distance between these distant phyla. Nevertheless, comparative analyses of genomes from probiotic bacteria revealed a core genome (probiogenome), which encodes key functions of this group of microorganisms.
Abstract
The human body is colonized by an enormous population of bacteria (microbiota) that provides the host with coding capacity and metabolic activities. Among the human gut microbiota are health-promoting indigenous species (probiotic bacteria) that are commonly consumed as live dietary supplements. Recent genomics-based studies (probiogenomics) are starting to provide insights into how probiotic bacteria sense and adapt to the gastrointestinal tract environment. In this Review, we discuss the application of probiogenomics in the elucidation of the molecular basis of probiosis using the well-recognized model probiotic bacteria genera Bifidobacterium and Lactobacillus as examples.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). This article describes the bacterial diversity that occurs in the human gut, assessed using 16S rRNA gene-based libraries.
Seksik, P. et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52, 237–242 (2003).
Turroni, F., Ribbera, A., Foroni, E., van Sinderen, D. & Ventura, M. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94, 35–50 (2008).
Rajilic-Stojanovic, M., Smidt, H. & de Vos, W. M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136 (2007). This review provides an integrated summary of data from culture-independent studies of the human gut microbiota.
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Guarner, F. & Malagelada, J. R. Gut flora in health and disease. Lancet 361, 512–519 (2003).
Hooper, L. V. & Gordon, J. I. Commensal host-bacterial relationships in the gut. Science 292, 1115–1118 (2001).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Samuel, B. S. & Gordon, J. I. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc. Natl Acad. Sci. USA 103, 10011–10016 (2006).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Kassinen, A. et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133, 24–33 (2007).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006). References 13 and 14 provide evidence for significant microbiota alterations in functional bowel disorders.
Food and Agriculture Organization of the United Nations and World Health Organization. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. (FAO/WHO, Cordoba, Argentina, 2001).
Marco, M. L., Pavan, S. & Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17, 204–210 (2006).
O'Hara, A. M. & Shanahan, F. Mechanisms of action of probiotics in intestinal diseases. Scientific World J. 7, 31–46 (2007).
Saxelin, M., Tynkkynen, S., Mattila-Sandholm, T. & de Vos, W. M. Probiotic and other functional microbes: from markets to mechanisms. Curr. Opin. Biotechnol. 16, 204–211 (2005).
Ventura, M. et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007).
Joyce, A. R. & Palsson, B. O. The model organism as a system: integrating 'omics' data sets. Nature Rev. Mol. Cell Biol. 7, 198–210 (2006).
Ventura, M. et al. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56, 2783–2792 (2006).
Tissier, M. H. Recherche Sur La Flore Intestinale Des Nourissons (Etat Normal Et Pathologique). Thesis, Univ. Paris, France (1906).
Ventura, M., Canchaya, C., Fitzgerald, G. F., Gupta, R. S. & van Sinderen, D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91, 351–372 (2007).
Schell, M. A. et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl Acad. Sci. USA 99, 14422–14427 (2002).
Gibson, G. R. & Roberfroid, M. B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412 (1995).
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R. & White, B. A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev. Microbiol. 6, 121–131 (2008).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T. & Gordon, J. I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl Acad. Sci. USA 96, 9833–9838 (1999).
Hinz, S. W., Verhoef, R., Schols, H. A., Vincken, J. P. & Voragen, A. G. Type I arabinogalactan contains β-D-Galp-(1→3)-β-D-Galp structural elements. Carbohydr. Res. 340, 2135–2143 (2005).
Ryan, S. M., Fitzgerald, G. F. & van Sinderen, D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72, 5289–5296 (2006).
Maze, A., O' Connell-Motherway, M., Fitzgerald, G. F., Deutscher, J. & van Sinderen, D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73, 545–553 (2007).
van den Broek, L. A., Hinz, S. W., Beldman, G., Vincken, J. P. & Voragen, A. G. Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 52, 146–163 (2008). This paper provides the most up-to-date description of the enzymes encoded by bifidobacteria that are involved in the hydrolysis of carbohydrates.
Siezen, R. et al. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific Gram-positive bacteria. BMC Genomics 7, 126 (2006).
Hooper, L. V., Midtvedt, T. & Gordon, J. I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 (2002).
Hoskins, L. C. et al. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J. Clin. Invest. 75, 944–953 (1985).
Ruas-Madiedo, P., Gueimonde, M., Fernandez-Garcia, M., de los Reyes-Gavilan, C. G. & Margolles, A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 74, 1936–1940 (2008).
Ventura, M., van Sinderen, D., Fitzgerald, G. F. & Zink, R. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek 86, 205–223 (2004).
Ehrmann, M. A., Korakli, M. & Vogel, R. F. Identification of the gene for β-fructofuranosidase of Bifidobacterium lactis DSM10140(T) and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46, 391–397 (2003).
Katayama, T. et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186, 4885–4893 (2004).
Ryan, S. M., Fitzgerald, G. F. & van Sinderen, D. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71, 3475–3482 (2005).
Gonzalez, R., Klaassens, E. S., Malinen, E., de Vos, W. M. & Vaughan, E. E. Differential transcriptional response of Bifidobacterium longum to human milk, formula milk and galactooligosaccharide. Appl. Environ. Microbiol. 74, 4686–4694 (2008).
Liepke, C. et al. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur. J. Biochem. 269, 712–718 (2002).
Ivanov, D. et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281, 17246–17252 (2006).
Potempa, J., Korzus, E. & Travis, J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J. Biol. Chem. 269, 15957–15960 (1994).
Sonnenburg, J. L., Chen, C. T. & Gordon, J. I. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4, e413 (2006). This paper describes the crosstalk that exists between bifidobacteria and Bacteroides in the murine intestine as well as between these bacteria and their hosts.
Kato, S., Haruta, S., Cui, Z. J., Ishii, M. & Igarashi, Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl. Environ. Microbiol. 71, 7099–7106 (2005).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Klijn, A., Mercenier, A. & Arigoni, F. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29, 491–509 (2005).
Aas, J. A. et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46, 1407–1417 (2008).
Makarova, K. S. & Koonin, E. V. Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208 (2007).
Claesson, M. J. et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl Acad. Sci. USA 103, 6718–6723 (2006).
Pfeiler, E. A. & Klaenhammer, T. R. The genomics of lactic acid bacteria. Trends Microbiol. 15, 546–553 (2007).
van de Guchte, M. et al. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl Acad. Sci. USA 103, 9274–9279 (2006).
Callanan, M. et al. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190, 727–735 (2008).
Altermann, E. et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl Acad. Sci. USA 102, 3906–3912 (2005).
Walter, J. et al. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69, 2044–2051 (2003).
Bron, P. A., Grangette, C., Mercenier, A., de Vos, W. M. & Kleerebezem, M. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186, 5721–5729 (2004). This manuscript provides insight into the interactions between a commensal bacterium and its murine host.
Oozeer, R. et al. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 71, 1356–1363 (2005).
Denou, E. et al. Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 189, 8109–8119 (2007).
Denou, E. et al. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190, 3161–3168 (2008).
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Tannock, G. W. et al. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66, 2578–2588 (2000).
Martin, F. P. et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4, 157 (2008).
Hickson, M. et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. Brit. Med. J. 335, 80 (2007).
Sullivan, A. & Nord., C. E. Probiotics and gastrointestinal diseases. J. Intern. Med. 257, 78–92 (2005).
Kelly, M. C., Mequio, M. J. & Pybus, V. Inhibition of vaginal lactobacilli by a bacteriocin-like inhibitor produced by Enterococcus faecium 62–66: potential significance for bacterial vaginosis. Infect. Dis. Obstet. Gynecol. 11, 147–156 (2003).
Corr, S. C. et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl Acad. Sci. USA 104, 7617–7621 (2007). This study identified the first molecular mechanism by which probiotic bacteria modulate the microbiota in vivo.
Casey, P. G. et al. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 73, 1858–1863 (2007).
Makarova, K. et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA 103, 15611–15616 (2006). This landmark study provided a large tranche of genomic data to allow studies of genome evolution in lactic acid bacteria.
Kleerebezem, M. et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl Acad. Sci. USA 100, 1990–1995 (2003). This is the first article describing the genome sequence of a member of the genus Lactobacillus.
Pridmore, R. D. et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl Acad. Sci. USA 101, 2512–2517 (2004). This paper describes the genome of a commonly used probiotic bacterium belonging to the genus Lactobacillus.
Talarico, T. L., Casas, I. A., Chung, T. C. & Dobrogosz, W. J. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 32, 1854–1858 (1988).
Santos, F. et al. The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiology 154, 81–93 (2008).
Sriramulu, D. D. et al. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J. Bacteriol. 190, 4559–4567 (2008).
Morita, H. et al. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15, 151–161 (2008).
Euzeby, J. P. List of bacterial names with standing in nomenclature: a folder available on the internet. Int. J. Syst. Bacteriol. 47, 590–592 (1997).
Boekhorst, J. et al. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150, 3601–3611 (2004).
Berger, B. et al. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 189, 1311–1321 (2007).
Nicolas, P., Bessieres, P., Ehrlich, S. D., Maguin, E. & van de Guchte, M. Extensive horizontal transfer of core genome genes between two Lactobacillus species found in the gastrointestinal tract. BMC Evol. Biol. 7, 141 (2007).
Klaenhammer, T. R., Barrangou, R., Buck, B. L., Azcarate-Peril, M. A. & Altermann, E. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29, 393–409 (2005).
Canchaya, C., Claesson, M. J., Fitzgerald, G. F., van Sinderen, D. & O'Toole, P. W. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152, 3185–3196 (2006).
Makarova, K. et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA 103, 15611–15616 (2006).
Claesson M. J., von Sinderen, D. & O'Toole, P. W. Lactobacillus phylogenomics — towards a reclassification of the genus. Int. J. Sys. Evo. Microbiol. (in press).
Teuber, M., Meile, L. & Schwarz, F. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek 76, 115–137 (1999).
Koonin, E. V., Makarova, K. S. & Aravind, L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742 (2001).
Lloyd, A. L., Rasko, D. A. & Mobley, H. L. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189, 3532–3546 (2007).
Pretzer, G. et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187, 6128–6136 (2005).
Grangette, C. et al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl Acad. Sci. USA 102, 10321–10326 (2005).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). This paper describes the bacterial diversity that exists in the gut of numerous mammals.
Lee, J. H. et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9, 247 (2008).
Leahy, S. C., Higgins, D. G., Fitzgerald, G. F. & van Sinderen, D. Getting better with bifidobacteria. J. Appl. Microbiol. 98, 1303–1315 (2005).
Acknowledgements
Work in the laboratories of D.v.S. and P.W.O.T. is supported by a Science Foundation Ireland Centres for Science, Engineering & Technology (SFI CSET) award to the Alimentary Pharmabiotic Centre and a Department of Agriculture and Food (DAF)/Health Research Board, Food-Health Research Initiative (HRB FHRI) FHRI award to the ELDERMET project. M.V. was supported by an Italian Award for Outstanding Young Researcher scheme “Incentivazione alla mobilità di studiosi stranieri e italiani residente all'estero” 2005–2009, a Marie Curie Reintegration Grant (MERG-CT-2005-03,080) and Parmalat spa, Italy. We also thank C. Canchaya for helpful discussions. Work on genomics of lactobacilli at North Carolina State University, USA, is supported by the NC Dairy Foundation, Danisco USA Inc. and Dairy Management Inc.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Microbiota
-
The collective microbial community or population that resides in a particular locale at a given time.
- Phylotypes
-
Groups of bacteria that are defined by percentage identity in their 16S rRNA gene sequences.
- Neighbour-joining tree
-
A tree that reconstructs the evolutionary development of organisms on the basis of distances between pairs of taxa.
- Omics
-
The integration of genomics methodology and data with functional genomic analyses involving transcriptomics, proteomics, metabolomics and interactomics.
- Prebiotics
-
Growth substrates that are preferentially (or ideally, exclusively) metabolized by a single genus or species and that may thus be used as dietary supplements to promote growth of a targeted health-promoting microorganism.
- Transcriptome
-
The subset of genes that are transcribed in an organism. It represents dynamic links between a genome, proteins and cellular phenotypes.
- Synteny
-
Genetic linkage or conservation of gene order.
- Bacteriocins
-
Proteinaceous substances that are produced by one bacterium to kill another bacterium, usually by inducing leakage or lysis. Bacteriocins are composed of one or two short peptides that can be post-translationally modified.
- COGs
-
Clusters of orthologous groups are delineated by comparing protein sequences that are encoded in complete genomes, representing major phylogenetic lineages. Each COG consists of individual proteins or groups of paralogues from at least 3 lineages and thus corresponds to an ancient conserved domain.
- Autochthonous
-
Members of the microbiota that are growing where they are found, as distinct from transient species that are only passing through the environment.
- Pseudoparalogous
-
An extra copy of a gene that is already present in a genome that was acquired by lateral gene transfer rather than by gene duplication.
- Microbiome
-
The collective genome of microbial communities.
Rights and permissions
About this article
Cite this article
Ventura, M., O'Flaherty, S., Claesson, M. et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol 7, 61–71 (2009). https://doi.org/10.1038/nrmicro2047
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2047
This article is cited by
-
NMGMDA: a computational model for predicting potential microbe–drug associations based on minimize matrix nuclear norm and graph attention network
Scientific Reports (2024)
-
The gut-lung axis in critical illness: microbiome composition as a predictor of mortality at day 28 in mechanically ventilated patients
BMC Microbiology (2023)
-
GACNNMDA: a computational model for predicting potential human microbe-drug associations based on graph attention network and CNN-based classifier
BMC Bioinformatics (2023)
-
A novel microbe-drug association prediction model based on stacked autoencoder with multi-head attention mechanism
Scientific Reports (2023)
-
Probiogenomics of Leuconostoc Mesenteroides Strains F-21 and F-22 Isolated from Human Breast Milk Reveal Beneficial Properties
Probiotics and Antimicrobial Proteins (2023)