Key Points
-
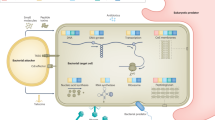
Bacteria produce a large variety of antimicrobial compounds directed against other bacterial species competing for the same resources. To mount an effective attack on their competitors some bacteria coordinate the production of toxins through intercellular communication with secreted species or strain-specific signalling molecules. To avoid suicide, producers also coordinate the expression of immunity proteins to protect themselves against their own toxins. Recently it has been discovered that in some cases subpopulations of isogenic bacteria can direct their weaponry against their own siblings. This phenomenon occurs in Streptococcus pneumoniae during natural transformation and in starving cells of Bacillus subtilis, and has been termed fratricide and cannibalism, respectively.
-
Cannibalism takes place during the early stages of sporulation in B. subtilis, when the bacterial population suffer from severe nutritional stress. Under such conditions, cells that have reached a certain stage in their sporulation differentiation process launch a coordinated attack on their siblings by secreting bacteriocin-like toxins. The targeted cells have not reached the same level of differentiation as their attackers, and therefore do not express the immunity proteins needed to fend off the attack. This behaviour was termed cannibalism as it causes the release of nutrients that the attacking cells can feed on to avoid, or at least delay, entry into sporulation.
-
Fratricide takes place in mixed populations of competent and non-competent pneumococci. Competent pneumococci secrete toxins that lyse their non-competent sister cells, liberating DNA and other cell constituents. This DNA-release mechanism is believed to increase the efficiency of gene exchange between pneumococci and other closely related streptococcal species under natural conditions. Evidence indicates that fratricide also contributes to virulence during human infections, as it mediates the release of the important virulence factor pneumolysin.
-
Cannibalism and fratricide represent interesting examples of a new kind of collaborative behaviour in bacteria. In these cases, some members of a population of isogenic bacteria (a subgroup), collaborate to attack their siblings. Previously, such collaborative behaviour has only been described for 'chemical warfare' between genetically non-identical bacteria.
Abstract
Cannibalism and fratricide refer to the killing of genetically identical cells (siblings) that was recently documented in two Gram-positive species, Bacillus subtilis and Streptococcus pneumoniae, respectively. Cannibalism occurs during the early stages of sporulation in B. subtilis, whereas fratricide occurs in S. pneumoniae during natural genetic transformation. Here, we compare and contrast these two phenomena and discuss whether these processes are fundamentally different from the more traditional 'chemical warfare' among bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Eijsink, V. G. et al. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Van Leeuwenhoek 81, 639–654 (2002).
Czaran, T. L., Hoekstra, R. F. & Pagie, L. Chemical warfare between microbes promotes biodiversity. Proc. Natl Acad. Sci. USA 99, 786–790 (2002).
Kleerebezem, M. & Quadri, L. E. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22, 1579–1596 (2001).
González-Pastor, J. E., Hobbs, E. C. & Losick, R. Cannibalism by sporulating bacteria. Science 301, 510–513 (2003). The first study to describe cannibalism in cultures of sporulating B. subtilis cells.
Ellermeier, C. D., Hobbs, E. C., Gonzalez-Pastor, J. E. & Losick, R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124, 549–559 (2006). Describes the elucidation of a three-protein signal-transduction pathway that controls immunity to one of the two toxins involved in cannibalism.
Guiral, S., Mitchell, T. J., Martin, B. & Claverys, J. P. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl Acad. Sci. USA 102, 8710–8715 (2005). This paper reports that competent pneumococci lyse their non-competent siblings on agar plates. It identifies a two-peptide bacteriocin, CibAB, and a putative murein hydrolase, CbpD, required for this phenomenon, termed allolysis, which elicits the release of the major virulence factor pneumolysin.
Håvarstein, L. S., Martin, B., Johnsborg, O., Granadel, C. & Claverys, J. P. New insights into pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59, 1297–1307 (2006). Identifies a competence-induced immunity protein, ComM, that protects competent cells from the action of lytic enzymes, two of which are competence induced (CbpD and LytA), and one of which is constitutively expressed (the LytC lysozyme).
Piggot, P. J. & Hilbert, D. W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7, 579–586 (2004).
Molle, V. et al. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50, 1683–1701 (2003).
Burbulys, D., Trach, K. A. & Hoch, J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64, 545–552 (1991). Elucidates the phosphorelay responsible for the control of Spo0A activity.
Fujita, M., Gonzalez-Pastor, J. E. & Losick, R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187, 1357–1368 (2005). This paper shows that sporulation requires high levels of Spo0A, and suggests that the genes involved in cannibalism are activated earlier and at a lower dose of Spo0A than the genes that have a direct role in spore formation.
Jiang, M., Shao, W., Perego, M. & Hoch, J. A. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38, 535–542 (2000).
Fujita, M. & Losick, R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19, 2236–2244 (2005).
Grossman, A. D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29, 477–508 (1995).
Errington, J. Regulation of endospore formation in Bacillus subtilis. Nature Rev. Microbiol. 1, 117–126 (2003). A comprehensive review of the regulation and molecular basis of spore formation in B. subtilis.
Claverys, J. P., Prudhomme, M., Mortier-Barrière, I. & Martin, B. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35, 251–259 (2000).
Claverys, J. P. & Håvarstein, L. S. Extra-cellular peptide control of competence for genetic transformation in Streptococcus pneumoniae. Frontiers Biosci. 7, 1798–1814 (2002). Reviews the regulation of competence development in Streptococcus pneumoniae , and discusses the role of cell–cell communication in this process.
Claverys, J. P., Prudhomme, M. & Martin, B. Induction of competence regulons as general stress responses in Gram-positive bacteria. Annu. Rev. Microbiol. 60, 451–475 (2006).
Prudhomme, M., Attaiech, L., Sanchez, G., Martin, B. & Claverys, J. P. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92 (2006).
Perego, M. in Cell–Cell Signaling in Bacteria (eds Dunny, G. M. & Winans, S. C.) 243–258 (ASM Press, Washington DC, 1999).
Perego, M. & Brannigan, J. A. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22, 1541–1547 (2001).
Dagkessamanskaia, A. et al. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51, 1071–1086 (2004).
Peterson, S. et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae. Mol. Microbiol. 51, 1051–1070 (2004).
Guiral, S., Dagkessamanskaia, A., Moscoso, M. & Claverys, J. P. in The Molecular Biology of Streptococci (eds Hakenbeck, R. M. & Chhatwal, G. S.) (Horizon Scientific, in the press).
Ween, O., Gaustad, P. & Håvarstein, L. S. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33, 817–827 (1999).
Luo, P. & Morrison, D. A. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185, 349–358 (2003).
Lee, M. S. & Morrison, D. A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181, 5004–5016 (1999).
Luo, P., Li, H. & Morrison, D. A. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50, 623–633 (2003).
Chen, G., Kumar, A., Wyman, T. H. & Moran, C. P. Jr. Spo0A-dependent activation of an extended10 region promoter in Bacillus subtilis. J. Bacteriol. 188, 1411–1418 (2006).
Kawulka, K. E. et al. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to α-carbon cross-links: formation and reduction of α-thio-α-amino acid derivatives. Biochemistry 43, 3385–3395 (2004).
Zheng, G., Yan, L. Z., Vederas, J. C. & Zuber, P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181, 7346–7355 (1999).
García, P., García, J. L., García, E. & López, R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene 43, 265–272 (1986).
García, P., González, M. P., García, E., García, J. L. & López, R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33, 128–138 (1999).
Steinmoen, H., Knutsen, E. & Håvarstein, L. S. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl Acad. Sci. USA 99, 7681–7686 (2002). Reports that cell lysis that affects a sub-population takes place in a competent culture of pneumococci, resulting in the release of cytoplasmic β-galactosidase and DNA. Shows that competence development and cell lysis are co-regulated, and that so-called choline-binding proteins are involved.
Steinmoen, H., Teigen, A. & Håvarstein, L. S. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J. Bacteriol. 185, 7176–7183 (2003). Shows that competent pneumococci can kill and lyse their non-competent siblings in mixed liquid cultures.
Moscoso, M. & Claverys, J. P. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 54, 783–794 (2004). Demonstrates the occurrence of competence-induced lytic events that lead to DNA release, that last for hours after competence induction and that are abolished in mutants lacking both the major autolysin LytA and the autolytic lysozyme LytC.
Kausmally, L., Johnsborg, O., Lunde, M., Knutsen, E. & Håvarstein, L. S. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187, 4338–4345 (2005).
van Belkum, M. J. et al. The bacteriocin lactococcin A specifically increases permeability of lactococcal cytoplasmic membranes in a voltage-independent, protein-mediated manner. J. Bacteriol. 173, 7934–7941 (1991).
Martínez-Cuesta, M. C. et al. Requirement of autolytic activity for bacteriocin-induced lysis. Appl. Environ. Microbiol. 66, 3174–3179 (2000).
Pei, J. & Grishin, N. V. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26, 275–277 (2001).
Stragier, P. To kill but not be killed: a delicate balance. Cell 124, 461–463 (2006).
Alloing, G., Martin, B., Granadel, C. & Claverys, J. P. Development of competence in Streptococcus pneumoniae: pheromone auto-induction and control of quorum-sensing by the oligopeptide permease. Mol. Microbiol. 29, 75–84 (1998).
Sung, C. K. & Morrison, D. A. Two distinct functions of ComW in stabilization and activation of the alternative σ factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187, 3052–3061 (2005).
Chung, J. D., Stephanopoulos, G., Ireton, K. & Grossman, A. D. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 176, 1977–1984 (1994).
Veening, J. W., Hamoen, L. W. & Kuipers, O. P. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 56, 1481–1494 (2005).
Dubnau, D. & Losick, R. Bistability in bacteria. Mol. Microbiol. 61, 564–572 (2006). This review discusses bistability, a random mechanism that switches on different genetic programmes within identical bacteria grown under the same conditions.
Veening, J. W., Smits, W. K., Hamoen, L. W. & Kuipers, O. P. Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J. Appl. Microbiol. 101, 531–541 (2006).
Lin, D., Qu, L. J., Gu, H. & Chen, Z. A 3.1-kb genomic fragment of Bacillus subtilis encodes the protein inhibiting growth of Xanthomonas oryzae pv. oryzae. J. Appl. Microbiol. 91, 1044–1050 (2001). Identifies the Skf killing factor (annotated as Ybc) as being active against the Gram-negative plant pathogen Xanthomonas oryzae , which might indicate that it is a broad spectrum bacteriocin.
Maamar, H. & Dubnau, D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56, 615–624 (2005).
Smits, W. K. et al. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56, 604–614 (2005).
Engelberg-Kulka, H. & Hazan, R. Cannibals defy starvation and avoid sporulation. Science 301, 467–468 (2003).
Lewis, K. Programmed death in bacteria. Microb. Mol. Biol. Rev. 64, 503–514 (2000).
Smits, W. K., Kuipers, O. P. & Veening, J. W. Phenotypic variation in bacteria: the role of feedback regulation. Nature Rev. Microbiol. 4, 259–271 (2006). An enjoyable review that discusses the molecular mechanisms of some of the natural bistable systems that have been identified in bacteria.
Wireman, J. W. & Dworkin, M. Morphogenesis and developmental interactions in myxobacteria. Science 189, 516–523 (1975).
Branda, S. S., Gonzalez-Pastor, J. E., Ben-Yehuda, S., Losick, R. & Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001).
Beiter, K. et al. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16, 401–407 (2006).
Buchanan, J. T. et al. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16, 396–400 (2006).
Bergmann, S., Rohde, M. & Hammerschmidt, S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72, 2416–2419 (2004).
Bergmann, S., Rohde, M., Chhatwal, G. S. & Hammerschmidt, S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40, 1273–1287 (2001).
Hamoen, L. W., Venema, G. & Kuipers, O. P. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149, 9–17 (2003).
Philips, Z. E. V. & Strauch, M. A. Bacillus subtilis sporulation and stationary phase expression. Cell. Mol. Life Sci. 59, 392–402 (2002).
Cotter, P. D., Hill, C. & Ross, R. P. Bacteriocins: developing innate immunity for food. Nature Rev. Microbiol. 3, 777–788 (2005).
Aizenman, E., Engelberg-Kulka, H. & Glaser, G. An Escherichia coli chromosomal 'addiction module' regulated by guanosine 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA 93, 6059–6063 (1996).
Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. & Hazan, R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS. Genet. 2, e135 (2006).
Gerdes, K., Christensen, S. K. & Lobner-Olesen, A. Prokaryotic toxin–antitoxin stress response loci. Nature Rev. Microbiol. 3, 371–382 (2005).
Nieto, C. et al. The chromosomal relBE2 toxin–antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin–antitoxin interaction. Mol. Microbiol. 59, 1280–1296 (2006).
Nieto, C. et al. The yefM-yoeB toxin–antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J. Bacteriol. 27 Oct 2006 (doi: 10.1128/JB.01130-06)
Pandey, D. P. & Gerdes, K. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 (2005).
Pedersen, K., Christensen, S. K. & Gerdes, K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45, 501–510 (2002).
Pozzi, G. et al. Competence for genetic transformation in Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178, 6087–6090 (1996).
Ramirez, M., Morrison, D. A. & Tomasz, A. Ubiquitous distribution of the competence related genes comA and comC among isolates of Streptococcus pneumoniae. Microb. Drug Res. 3, 39–52 (1997).
Whatmore, A. M., Barcus, V. A. & Dowson, C. G. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181, 3144–3154 (1999).
Håvarstein, L. S., Hakenbeck, R. & Gaustad, P. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179, 6589–6594 (1997).
Tortosa, P. & Dubnau, D. Competence for transformation: a matter of taste. Curr. Opin. Microbiol. 2, 588–592 (1999).
Acknowledgements
We would like to thank present and past members of the two laboratories for their contributions to the work discussed in this review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Bacteriocin
-
A bacterially produced, small, heat-stable peptide that is active against other bacteria and to which the producer has a specific immunity mechanism. Can be narrow or broad spectrum.
- Transformation
-
The uptake and incorporation of exogenous 'naked' DNA directly from the environment.
- Response regulator
-
A bacterial gene-regulatory protein that controls gene expression in response to external signals. Most response regulators consist of two domains: a DNA-binding domain and a regulatory domain, the activity of which is modulated (indirectly) by the external signal.
- Phosphorelay protein
-
A protein involved in a complex pathway in which phosphoryl groups are transferred through several signal-transduction proteins before reaching the target protein.
- Krebs cycle
-
Also known as the tricarboxylic acid (or TCA) cycle. A metabolic pathway that breaks down the products of carbohydrate, fat and protein metabolism into carbon dioxide and water to generate energy. It also provides precursors for other compounds, such as certain amino acids.
- Autocrine
-
The activation of cellular receptors by ligands produced by the same cell.
- Two-component regulatory system
-
A protein pair involved in signal transduction in which the sensor is a histidine kinase, the effector is a response regulator and the signalling is based on the phosphotransfer between these two components.
- Alternative σ factor
-
An alternative σ factor is produced under specific conditions and allows the RNA polymerase to transcribe a different set of genes from the housekeeping σ factor, σ70.
- Type II CAAX prenyl endopeptidase
-
A protein belonging to a superfamily of putative membrane-bound metalloproteases.
- Lanthionine
-
Lanthionine is formed when a dehydrated serine or threonine is covalently bridged (through the sulphur atom) with a cysteine.
- Bistable switch
-
A random mechanism that switches on different genetic programmes in identical bacteria that are grown under the same conditions.
- Toxin–antitoxin
-
Paired loci found in the chromosomes of almost all free-living bacteria, and many plasmids and phage genomes. They encode a toxin and its antidote, which have been shown to contribute to plasmid stability by a mechanism called post-segregational killing. They are also proposed to function in bacterial programmed cell death or stress physiology.
- Pherotype
-
Streptococci that produce identical or cross-inducing competence pheromones (CSPs) belong to the same pherotype.
Rights and permissions
About this article
Cite this article
Claverys, JP., Håvarstein, L. Cannibalism and fratricide: mechanisms and raisons d'être. Nat Rev Microbiol 5, 219–229 (2007). https://doi.org/10.1038/nrmicro1613
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1613
This article is cited by
-
Bicyclostreptins are radical SAM enzyme-modified peptides with unique cyclization motifs
Nature Chemical Biology (2022)
-
Impact of previous macrolide use on invasive pneumococcal disease due to erythromycin-resistant serotypes in adults over 59 years of age
European Journal of Clinical Microbiology & Infectious Diseases (2022)
-
Toward understanding the signals of bacteriocin production by Streptococcus spp. and their importance in current applications
World Journal of Microbiology and Biotechnology (2021)
-
Bacterial bug-out bags: outer membrane vesicles and their proteins and functions
Journal of Microbiology (2020)
-
Inhibitory effect of streptococci on the growth of M. catarrhalis strains and the diversity of putative bacteriocin-like gene loci in the genomes of S. pneumoniae and its relatives
AMB Express (2017)