Key Points
-
Bacteriophages (phages) can devastate bacterial strains that are used in fermentations and bioprocesses.
-
Comparative genomic analyses can be used to streamline the construction of genetic systems that are designed to protect bioprocessing strains against phage attack.
-
Gene-silencing techniques, such as antisense-RNA targeting of essential phage-encoded genes (for example, DNA replication) can effectively inhibit the propagation of virulent phages.
-
Trans-dominant negative mutant proteins that are derived from phage-encoded genes can be used to sabotage the function of multimeric phage–protein complexes by a process called subunit poisoning.
-
The native role of some phage-encoded genes is to protect lysogens against superinfecting phages. Such genes can also be exploited to protect non-lysogenic bioprocessing cultures from superinfection (for example, superinfection exclusion or immunity).
-
Selected engineered systems have been composed of phage-encoded cis regulatory elements, such as phage origins of DNA replication or phage promoters that drive abortive or suicide systems in the infected cell (for example, phage-triggered suicide systems).
Abstract
Bacteriophages (phages) have the potential to interfere with any industry that produces bacteria as an end product or uses them as biocatalysts in the production of fermented products or bioactive molecules. Using microorganisms that drive food bioprocesses as an example, this review will describe a set of genetic tools that are useful in the engineering of customized phage-defence systems. Special focus will be given to the power of comparative genomics as a means of streamlining target selection, providing more widespread phage protection, and increasing the longevity of these industrially important bacteria in the bioprocessing environment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pasteur, L. Mémoire sur la fermentation appelée lactique. C. R. Acad. Sci. 45, 913–916 (1857).
Lister, J. In Milestones in Microbiology: 1556 to 1940 (ed. Brock, T.D.) 58 (American Society for Microbiology Press, Washington DC, 1998).
Mundt, O. The lactic acid streptococci. In Bergey's Manual of Systematic Bacteriology (ed. Sneath, P. H. A., Mair, N. S., Sharpe, M. E. & Holt, J. G.) 2, 1065–1066 (Williams & Wilkins, Baltimore 1986).
Löhnis, F. Die Benennung der Milchsaürebakterien. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 22, 553–555 (1909).
Schleifer, K. H. et al. Transfer of Streptococcus lactis and related species to the genus Lactococcus. Syst. Appl. Microbiol. 6, 183–195 (1985).
Thunell, R. K. & Sandine, W. E. In Bacterial Starter Cultures for Foods (ed. Gilland, S. E.) 127–144 (CRC Press, BocaRaton, 1985).
Twort, F. An investigation on the nature of ultra-microscopic viruses. Lancet 2, 1241–1243 (1915).
D'Herelle, F. In Milestones in Microbiology: 1556 to 1940 (ed. Brock, T. D.) 157 (American Society for Microbiology Press, Washington DC, 1998).
Whitehead, H. R. & Cox, G. A. The occurrence of bacteriophage in cultures of lactic streptococci, a preliminary note. N. Z. J. Sci. Technol. 16, 319 (1935).
Bruttin, A. et al. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63, 3144–3150 (1997).
Moineau, S. et al. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J. Dairy Sci. 79, 2104–2111 (1996).
Brüssow, H., Bruttin, A., Desiere, F., Lucchini, S. & Foley, S. Molecular ecology and evolution of Streptococcus thermophilus bacteriophage — a review. Virus Genes 16, 95–109 (1998).
Binetti, A. G. & Reinheimer, J. A. Thermal and chemical inactivation of indigenous Streptococcus thermophilus bacteriophages isolated from Argentinean dairy plants. J. Food Prot. 63, 509–515 (2000).
Madera, C., Monjardin, C. & Suarez, J. E. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl. Environ. Microbiol. 70, 7365–7371 (2004).
Chopin, M. C. Resistance of 17 mesophilic lactic Streptococcus bacteriophages to pasteurization and spray drying. J. Dairy Res. 47, 131–139 (1980).
Hassan, A. N., Awad, S. & Muthukumarappan, K. Effects of exopolysaccharide-producing cultures on the viscoelastic properties of reduced-fat Cheddar cheese. J. Dairy Sci. 88, 4221–4227 (2005).
Tremblay, D. M. & Moineau, S. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255, 63–76 (1999).
Stanley, E., Fitzgerald, G. F., Le Marrec, M. C., Fayard, B. & van Sinderen, D. Sequence analysis and characterization of O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiol. 143, 3417–3429 (1997).
Lucchini, S., Desiere, F. & Brüssow, H. The structural gene module in Streptococcus thermophilus bacteriophage φSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology 246, 63–73 (1998).
Lucchini, S., Desiere, F. & Brüssow, H. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260, 232–243 (1999).
Stanley, E., Walsh, L., van der Zwet, A., Fizgerald, G. F. & van Sinderen, D. Identification of four loci isolated from two Streptococcus thermophilus phage genomes responsible for mediating bacteriophage resistance. FEMS Microbiol. Lett. 182, 271–277 (2000).
Lévesque, C. et al. Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 71, 4057–4068 (2005).
Brüssow, H. & Desiere, F. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39, 213–222 (2001).
Lucchini, S., Desiere, F. & Brüssow, H. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virology 73, 8647–8656 (1999).
Le Marrec, C. et al. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63, 3246–3253 (1997). Differentiates S. thermophilus phages into two distinct evolutionary lineages and describes a PCR-based diagnostic method for their classification in vitro.
Ventura, M., Bruttin, A., Canchaya, C. & Brüssow, H. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302, 21–32 (2002).
Ventura, M. et al. Transcription mapping as a tool in phage genomics: the case of the temperate Streptococcus thermophilus phage Sfi21. Virology 296, 62–76 (2002).
Ventura, M. & Brüssow, H. Temporal transcription map of the virulent Streptococcus thermophilus bacteriophage Sfi19. Appl. Environ. Microbiol. 70, 5041–5046 (2004).
Duplessis, M., Russell, W., Romero, D. & Moineau, S. Global gene expression analysis of two Streptococcus thermophilus bacteriophages using DNA microarray. Virology 340, 192–208 (2005). Describes the use of DNA microarrays to monitor S. thermophilus phage gene regulation over the course of an ongoing lytic infection.
Wang, I. N., Smith, D. L. & Young, R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799–825 (2000).
Sable, S. & Lortal, S. The lysins of bacteriophages infecting lactic acid bacteria. Appl. Microbiol. Biotechnol. 43, 1–6 (1995).
Beresford, T. P., Ward, L. J. & Jarvis, A. W. Temporally regulated transcriptional expression of the genomes of lactococcal bacteriophages c2 and sk1. Appl. Environ. Microbiol. 59, 3708–3712 (1993).
Brüssow, H., Probst, A., Fremont, M. & Sidoti, J. Distinct Streptococcus thermophilus bacteriophages share an extremely conserved DNA fragment. Virology 200, 854–857 (1994).
Sturino, J. M. & Klaenhammer, T. R. Expression of antisense RNA targeted against Streptococcus thermophilus bacteriophages. Appl. Environ. Microbiol. 68, 588–596 (2002).
Masse, E., Escorcia, F. E. & Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383 (2003).
Sturino, J. M. & Klaenhammer, T. R. Bacteriophage defencesystems and strategies for lactic acid bacteria. In Advances in Applied Microbiology Vol. 56 (eds Laskin, A. I., Bennett, J. W. & Gadd, G. M.) 331–378 (Academic Press, San Diego 2005). A more comprehensive review of bacteriophage-defence strategies and systems for microorganisms that are used in bioprocessing environments.
Sturino, J. M. & Klaenhammer, T. R. Antisense RNA targeting primase interferes with bacteriophage replication in Streptococcus thermophilus. Appl. Environ. Microbiol. 70, 1735–1743 (2004).
Djordjevic, G., Bojovic, B., Miladinov, N. & Topisirovic, L. Cloning and molecular analysis of promoter-like sequences isolated from the chromosomal DNA of Lactobacillus acidophilus ATCC 4356. Can. J. Microbiol. 43, 61–69 (1997).
de Vos, W. M. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46, 281–295 (1987).
Bull, J. J., Jacobson, A., Badgett, M. R. & Molineux, I. J. Viral escape from antisense RNA. Mol. Microbiol. 28, 835–846 (1998).
Keppel, F., Fayet, O. & Georgopoulos, C. Strategies of bacteriophage DNA replication. In The Bacteriophages Vol. 2 (ed. Calendar, R.) 145–264 (Plenum Press, New York, 1988).
Hill, C., Miller, L. A. & Klaenhammer, T. R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 172, 6419–6426 (1990).
Foley, S., Lucchini, S., Zwahlen, M. C. & Brüssow, H. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology 250, 377–387 (1998).
Lamothe, G. et al. Characterization of the cro-ori region of the Streptococcus thermophilus virulent bacteriophage DT1. Appl. Environ. Microbiol. 71, 1237–1246 (2005).
O'Sullivan, D. J., Hill, C. & Klaenhammer, T. R. Effect of increasing the copy number of bacteriophage origins of replication in trans, on incoming-phage proliferation. Appl. Environ. Microbiol. 59, 2449–2456 (1993).
McGrath, S., Fitzgerald, G. F. & van Sinderen, D. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67, 608–616 (2001).
Lawrence, J. G., Hendrix, R. & Casjens, S. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9, 535–540 (2001).
Brüssow, H. & Hendrix, R. W. Phage genomics: small is beautiful. Cell 108, 13–16 (2002).
Bruttin, A., Foley, S. & Brüssow, H. DNA-binding activity of the Streptococcus thermophilus phage Sfi21 repressor. Virology 303, 100–109 (2002).
Bruttin, A., Desiere, F., Lucchini, S., Foley, S. & Brüssow, H. Characterization of the lysogeny DNA module from the temperate Streptococcus thermophilus bacteriophage Sfi21. Virology 233, 136–148 (1997).
Djordjevic, G. M., O'Sullivan, D. J., Walker, S. A., Conkling, M. A. & Klaenhammer, T. R. A triggered-suicide system designed as a defence against bacteriophages. J. Bacteriol. 179, 6741–6748 (1997). Demonstrates the successful implementation of phage-triggered suicide systems as a method for phage defence.
Djordjevic, G. M. & Klaenhammer, T. R. Bacteriophage-triggered defence systems: phage adaptation and design improvements. Appl. Environ. Microbiol. 63, 4370–4376 (1997).
O'Sullivan, D. J., Zagula, K. & Klaenhammer, T. R. In vivo restriction by LlaI is encoded by three genes, arranged in an operon with llaIM, on the conjugative Lactococcus plasmid pTR2030. J. Bacteriol. 177, 134–143 (1995).
Herskowitz, I. Functional inactivation of genes by dominant negative mutation. Nature 329, 219–222 (1987).
Notarnicola, S. M., Park, K., Griffith, J. D. & Richardson, C. C. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 270, 20215–20224 (1995).
Durmaz, E., Madsen, S. M., Israelsen, H. & Klaenhammer, T. R. Lactococcus lactis lytic bacteriophages of the P335 group are inhibited by overexpression of a truncated CI repressor. J. Bacteriol. 184, 6532–6544 (2002).
Lucchini, S., Sidoti, J. & Brüssow, H. Broad-range bacteriophage resistance in Streptococcus thermophilus by insertional mutagenesis. Virology 275, 267–277 (2000). Illustrates the use of insertional mutagenesis to identify host-encoded factors that are essential for bacteriophage replication.
Maguin, E., Prevost, H., Ehrlich, S. D. & Gruss, A. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J. Bacteriol. 178, 931–935 (1996).
Garbutt, K. C., Kraus, J. & Geller, B. L. Bacteriophage resistance in Lactococcus lactis engineered by replacement of a gene for a bacteriophage receptor. J. Dairy Sci. 80, 1512–1519 (1997).
Pedersen, M. B., Jensen, P. R., Janzen, T. & Nilsson, D. Bacteriophage resistance of a thyA mutant of Lactococcus lactis blocked in DNA replication. Appl. Environ. Microbiol. 68, 3010–3023 (2002). Describes a novel approach for the construction of starter-culture strains with a nutritional deficiency that severely limits the DNA-replication ability of phages during their lytic developmental cycle. It is noteworthy that the approach is generally effective against all phages within two of the major species groups that infect lactococci.
Pedersen, M. B., Koebmann, B. J., Jensen, P. R. & Nilsson, D. Increasing acidification of nonreplicating Lactococcus lactis ΔthyA mutants by incorporating ATPase activity. Appl. Environ. Microbiol. 68, 5249–5257 (2002).
Cogan, T. M., Peitersen, N. & Sellars, R. L. Starter systems. In Bulletin of the International Dairy Federation: Practical Phage Control 263, 16–23 (International Dairy Federation, Brussels, 1991).
Durmaz, E. & Klaenhammer, T. R. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defences in a single Lactococcus lactis strain. Appl. Environ. Microbiol. 61, 1266–1273 (1995).
Sing, W. D. & Klaenhammer, T. R. A strategy for rotation of different bacteriophage defences in a lactococcal single-strain starter culture system. Appl. Environ. Microbiol. 59, 365–372 (1993).
Heap, H. A. & Lawrence, R. C. The selection of starter strains for cheese making. N. Z. J. Dairy Sci. Technol. 11, 16–53 (1976).
Huggins, A. R. Progress in dairy starter culture technology. Food Technol. 38, 41 (1984).
Klaenhammer, T. R. Interactions of bacteriophages with lactic streptococci. In Advances in Applied Microbiology Vol. 30 (ed. Laskin, A. I.) 1–29 (Academic Press, New York, 1984).
Viscardi, M., Capparelli, R. & Iannelli, D. Rapid selection of phage-resistant mutants in Streptococcus thermophilus by immunoselection and cell sorting. Int. J. Food Microbiol. 89, 223–231 (2003).
Viscardi, M. et al. Selection of bacteriophage-resistant mutants of Streptococcus thermophilus. J. Microbiol. Methods 55, 109–119 (2003).
Sanders, M. E., Leonhard, P. J., Sing, W. D. & Klaenhammer, T. R. Conjugal strategy for construction of fast-acid producing, bacteriophage resistant lactic streptococci for use in dairy fermentations. Appl. Environ. Microbiol. 52, 1001–1007 (1986).
Klaenhammer, T. R. & Fitzgerald, G. F. Bacteriophage and bacteriophage resistance. In Genetics and Biotechnology of Lactic Acid Bacteria (eds Gasson M. J. & de Vos, W. M.) 106–168 (Chapman and Hall, London, 1994).
Johansen, E. Genetic engineering, modification of bacteria. In Encyclopedia of Food Microbiology (eds Robinson, R., Batt, C. & Patel, P.) 917–912 (Academic Press, London, 1999). An excellent resource describing the technical and regulatory challenges that must be addressed when developing safe, genetically modified microorganisms that are ultimately intended for human consumption.
Husson-Kao, C., Mengaud, J., Gripon, J. C., Benbadis, L. & Chapot-Chartier, M. P. Characterization of Streptococcus thermophilus strains that undergo lysis under unfavourable environmental conditions. Int. J. Food Microbiol. 55, 209–213 (2000).
Husson-Kao, C. et al. The Streptococcus thermophilus autolytic phenotype results from a leaky prophage. Appl. Environ. Microbiol. 66, 558–565 (2000).
Stanley, E., Fitzgerald, G. F. & van Sinderen, D. Characterisation of Streptococcus thermophilus CNRZ1205 and its cured and re-lysogenised derivatives. FEMS Microbiol. Lett. 176, 503–510 (1999).
Dinsmore, P. & Klaenhammer, T. R. Bacteriophage resistance in Lactococcus. Mol. Biotechnol. 4, 297–314 (1995).
Hjalt, T. A & Wagner, E. G. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 23, 580–587 (1995).
Kolb, F. A. et al. Bulged residues promote the progression of a loop–loop interaction to a stable and inhibitory antisense–target RNA complex. Nucleic Acids Res. 29, 3145–3153 (2001).
Acknowledgements
General support for the research on engineered phage-defence systems at North Carolina State University has been provided by Danisco USA, Inc., the US Department of Agriculture/National Research Initiative Competitive Grants Programme and the North Carolina Dairy Foundation. The authors also thank Chr. Hansen Inc. for their support of J. Sturino during the preparation of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Organoleptic
-
Being, affecting or relating to qualities (such as taste, colour, odour and feel) of a substance that stimulate the sense organs.
- Backslopping
-
An artisanal practice whereby a small portion of a previous batch is used to inoculate subsequent batches. From the standpoints of safety and product consistency, backslopping is not recommended owing to the risk of transferring and enriching for pathogenic microorganisms or bacteriophages, respectively.
- Starter culture
-
Concentrated preparations of viable microorganisms (usually strains of lactic acid bacteria or yeasts) that are added to bioprocessing systems to mediate the bioconversion of the substrate in an accelerated and more reproducible manner when compared to spontaneous fermentations. These preparations are product-optimized and are generally either freeze-dried or stored frozen.
- Phage unrelated
-
Character of strains or species whereby the bacteria in question exhibit distinct phage-sensitivity profiles, meaning that they are attacked by different groups of phages.
- Holin
-
Small, membrane-spanning protein that accumulates in the cytosolic membrane during the lytic life cycle of a phage. Holin proteins act as a gateway to the cell wall for the phage-encoded endolysin.
- Endolysin
-
Phage-encoded muralytic enzyme that degrades bacterial cell-wall polymers. During the lytic life cycle, endolysin accumulates in the cytosol and can cross the cytosolic membrane by the cooperative action of the phage-encoded holin.
- Efficiency of plaquing
-
(EOP). Calculated by dividing the phage titre (in plaque forming units (pfu) per ml) on the test strain (phage-resistant) by the phage titre in pfu per ml on the parent strain (phage-sensitive indicator).
- Prophage
-
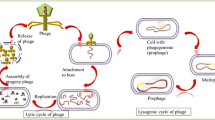
The latent form of a temperate bacteriophage in which its genome is integrated into the bacterial chromosome without causing disruption of the bacterial cell.
- Lysogen
-
A bacterium that contains a prophage integrated into its genome. Lysogens can induce the prophage into a lytic developmental cycle and cause cell lysis to release progeny phage. In the lysogen, the prophage remains quiescent and is effectively replicated once with every chromosomal division of the bacterium.
- Superinfection
-
Any phage infection that occurs after an earlier one; often describing a secondary infection of a lysogenic bacterium.
- Amber mutation
-
A nonsense mutation that introduces a premature UAG translational stop codon in a gene.
- Ochre mutation
-
A nonsense mutation that introduces a premature UAA translational stop codon in a gene.
- Phage out
-
The event whereby the starter culture inoculated in a bioprocessing system has been decimated by the lytic activity of phages to such a degree that the bioconversion is abandoned. Vats of partially cultured milk that have 'phaged out' are normally discarded.
- Phage-insensitive mutant
-
Phage-resistant mutants that are derived from a parent strain that are insensitive to the phage(s) in question. Phage-insentive mutants are generally the result of targeted phage-challenge assays, whereby a phage-sensitive parent strain is repeatedly challenged with a cocktail of two or more unrelated phages. This process is often referred to as phage hardening.
Rights and permissions
About this article
Cite this article
Sturino, J., Klaenhammer, T. Engineered bacteriophage-defence systems in bioprocessing. Nat Rev Microbiol 4, 395–404 (2006). https://doi.org/10.1038/nrmicro1393
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1393
This article is cited by
-
Strain and process engineering toward continuous industrial fermentation
Frontiers of Chemical Science and Engineering (2023)
-
Tn5 Transposon-based Mutagenesis for Engineering Phage-resistant Strains of Escherichia coli BL21 (DE3)
Journal of Microbiology (2023)
-
Application of CRISPR-Mediated Gene Editing for Crop Improvement
Molecular Biotechnology (2022)
-
Phage lysin that specifically eliminates Clostridium botulinum Group I cells
Scientific Reports (2020)
-
Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications
Cellular & Molecular Biology Letters (2016)