Key Points
-
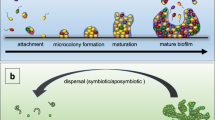
Bacterial biofilms can be considered to be an emergent form of bacterial life, in which communal life is completely different from bacteria that live as free-living cells.
-
Emergent properties of bacterial biofilms include social cooperation, resource capture and enhanced survival following exposure to antimicrobials, and cannot be understood and predicted through the study of free-living bacterial cells.
-
The physical scaffold of biofilm life is the matrix of extracellular polymeric substances (EPS) that keeps cells in the biofilm together and attaches them to substrata when colonizing surfaces. The matrix underlies the emergent properties of biofilms.
-
The emergent properties of the biofilm are the reason for the evolutionary success of biofilms and underlie the role of biofilms as global habitat formers.
Abstract
Bacterial biofilms are formed by communities that are embedded in a self-produced matrix of extracellular polymeric substances (EPS). Importantly, bacteria in biofilms exhibit a set of 'emergent properties' that differ substantially from free-living bacterial cells. In this Review, we consider the fundamental role of the biofilm matrix in establishing the emergent properties of biofilms, describing how the characteristic features of biofilms — such as social cooperation, resource capture and enhanced survival of exposure to antimicrobials — all rely on the structural and functional properties of the matrix. Finally, we highlight the value of an ecological perspective in the study of the emergent properties of biofilms, which enables an appreciation of the ecological success of biofilms as habitat formers and, more generally, as a bacterial lifestyle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vert, M. et al. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 84, 377–410 (2012).
Konopka, A. What is microbial community ecology? ISME J. 3, 1223–1230 (2009).
Stoodley, P., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
Ehrlich, H. L. & Newman, D. K. Geomicrobiology 5th edn (CRC press, 2008).
Meckenstock, R. et al. Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 49, 7073–7081 (2015).
Halan, B., Bühler, K. & Schmid, A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 30, 453–465 (2012).
De Vos, W. M. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes 1, 15005 (2015).
Costerton, J. W. et al. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464 (1987).
Shirtliff, M. & Leid, J. (eds) The Role of Biofilms in Device-Related Infections (Springer, 2009).
Flemming, H.-C. in Biofilm Highlights (eds Flemming, H.-C., Wingender, J. & Szewzyk, U.) 81–109 (Springer, 2011).
Wingender, J. & Flemming, H.-C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health 214, 417–423 (2011).
Little, B. J. & Lee, J. S. Microbiologically influenced corrosion: an update. Int. Mat. Rev. 59, 384–393 (2014).
Balzer, M., Witt, N., Flemming, H.-C. & Wingender, J. Accumulation of fecal indicator bacteria in river biofilms. Water Sci. Technol. 61, 1105–1111 (2010).
Morgan-Sastume, F., Larsen, P., Nielsen, J. L. & Nielsen, P. H. Characterization of the loosely attached fraction of activated sludge bacteria. Water Res. 42, 843–854 (2008).
Singer, S. W. et al. Posttranslational modification and sequence variation of redox-active proteins correlate with biofilm life cycle in natural microbial communities. ISME J. 4, 1348–1409 (2010).
Corning, P. A. The re-emergence of “emergence”: a venerable concept in search of a theory. Complexity 7, 18–30 (2002).
Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Saville, R. M. et al. Energy-dependent stability of Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 193, 3257–3264 (2011).
Neu, T. R. & Lawrence, J. R. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 23, 233–242 (2014).
Serra, D. O., Richter, A. M., Klauck, G., Mika, F. & Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 195, 5540–5554 (2013). A paper that highlights the important and underestimated role of cellulose for biofilm architecture.
Birjiniuk, A. et al. Single particle tracking reveals spatial and dynamic organization of the Escherichia coli biofilm matrix. New J. Phys. 16, 085014 (2014).
Persat, A. et al.: The mechanical world of bacteria. Cell 161, 988–997 (2005).
Jennings, L. K. et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl Acad. Sci. USA 112, 11353–11358 (2015).
Devaraj, A., Justice, S. S., Bakaletz, L. O. & Goodman, S. D. DNABII proteins play a central role in UPEC biofilm structure. Mol. Microbiol. 96, 1119–1135 (2015).
Böckelmann, U. et al. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 262, 31–38 (2006).
Hobley, L., Harkins, C., MacPhee, C. E. & Stanley-Wall, N. R. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39, 649–669 (2015).
Karimi, A., Karig, D., Kumar, A. & Ardekani, A. M. Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab Chip 15, 23–42 (2015).
Wilking, J. N. et al. Liquid transport facilitated by channels in Bacillus subtilis. Proc. Natl Acad. Sci. USA 110, 848–852 (2013).
Patel, R. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 437, 41–47 (2005).
Lovley, D. R. & Malvankar, N. S. Seeing is believing: novel imaging techniques help clarify microbial nanowire structure and function. Environ. Microbiol, 17, 2209–2215 (2015).
Koch, C. et al. Coupling electric energy and biogas production in anaerobic digesters — impact on the microbiome. RSC Adv. 5, 31329–31340 (2015).
Secor, P. R. et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18, 1–11 (2015).
Schooling, S. R. & Beveridge, T. J. Membrane vesicles: an overlooked component of matrices of biofilms. J. Bacteriol. 188, 5945–5957 (2006).
Billings, N., Birjiniuk, A., Samad, T. S., Doyle, P. S. & Ribbeck, K. Material properties of biofilms — a review of methods for understanding permeability and mechanics. Rep. Prog. Phys. 78, 036601 (2015).
Raymond, J. & Alsop, E. B. Microbial evolution in extreme environments: microbial migration, genomic highways, and geochemical barriers in hydrothermal ecosystems. Environ. Syst. Res. 4, 14 (2015).
Van Gestel, J., Vlamakis, H. & Kolter, R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 13, e1002141 (2015).
Houry, A. et al. Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl Acad. Sci. USA 109, 13088–13093 (2012). A report of surprisingly fast tunnelling movement of some microorganisms in the biofilm matrix, and an investigation of the consequences of this movement.
Petrova, O. E. & Sauer, K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78 (2016).
Whitfield, G. B., Marmont, L. S. & Howell, P. L. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 6, 471 (2015).
Helm, R. F. & Potts, M. in Ecology of Cyanobacteria II: Their Diversity in Space and Time (ed Whitton, B. A.) 461–480 (Springer, 2012).
Flemming, H.-C. The perfect slime. Colloids Surf. B. Biointerfaces 86, 251–259 (2011).
Weaver, L., Webber, J. B., Hickson, A. C., Abraham, P. M. & Close, M. E. Biofilm resilience to desiccation in groundwater aquifers: a laboratory and field study. Sci. Total Environ. 514, 281–289 (2015).
Flemming, H.-C. Biodeterioration of synthetic materials — a brief review. Mat. Corr. 61, 986–992 (2010).
Flemming, H.-C. & Leis, A. in Encyclopedia of Environmental Microbiology Vol. 5 (ed. Bitton, G.) 2958–2967 (Wiley-Interscience, 2002).
Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M. & Packmann, A. I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 14, 251–263 (2016).
Métevier, R., Bourven, I., Labanowski, J. & Guibaud, G. Interaction of erythromycin ethylsuccinate and acetaminophen with protein fraction of extracellular polymeric substances (EPS) from various bacterial aggregates. Environ. Sci. Pollut. Res. Int. 20, 7275–7285 (2013).
Dobor, J., Varga, M. & Záray, G. Biofilm controlled sorption of selected acidic drugs on river sediments characterized by different organic carbon content. Chemosphere 87, 105–110 (2012).
Writer, J. H., Barber, L. B., Ryan, J. N. & Bradley, P. M. Biodegradation and attenuation of steroidal hormones and alkylphenols by stream biofilms and sediments. Environ. Sci. Technol. 45, 4370–4376 (2011).
Späth, R., Flemming, H.-C. & Wuertz, S. Sorption properties of biofilms. Water Sci. Technol. 37, 207–210 (1998).
Schmitt, J., Nivens, D., White, D. C. & Flemming, H.-C. Changes of biofilm properties in response to sorbed substances — an FTIR–ATR-study. Water Sci. Technol. 32, 149–155 (1995).
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisky, G. G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016).
López, D., Vlamakis, H. & Kolter, R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74, 609–618 (2009).
Pinchuk, G. E. et al. Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl. Environ. Microbiol. 74, 1198–1208 (2008).
Mulcahy, H., Charron-Mazenod, L. & Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12, 1621–1629 (2010).
Kaplan, J. B. in Microbial Biofilms: Methods and Protocols (ed Donelli, G.) 203–213 (Springer, 2014).
Zrelli, K. et al. Bacterial biofilm mechanical properties persist upon antibiotic treatment and survive cell death. New J. Phys. 15, 125026 (2013).
Decho, A. W., Visscher, P. T. & Reid, R. P. Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Paleo 219, 71–86 (2005).
Körstgens, V., Wingender, J., Flemming, H.-C. & Borchard, W. Influence of calcium ion concentration on the mechanical properties of a model biofilm of Pseudomonas aeruginosa. Water Sci. Technol. 43, 49–57 (2001).
Nielsen, P. H. & Jahn, A. in Microbial Extracellular Polymeric Substances (eds Wingender, J., Neu, T. R. & Flemming, H.-C.) 49–72 (Springer, 1999).
Oppenheimer-Shaanan, Y. et al. Spatio-temporal assembly of functional mineral scaffolds within microbial biofilms. NPJ Biofilms Microbiomes 2, 15031 (2016).
Decho, A. W. Overview of biopolymer-induced mineralization: what goes on in biofilms? Ecol. Eng. 36, 137–144 (2010).
Braun, B., Richert, I. & Szewzyk, U. Detection of iron-depositing Pedomicrobium species in native biofilms from the Odertal National Park by a new, specific FISH probe. J. Microbiol. Methods 79, 37–43 (2009).
Grumbein, S., Opitz, M. & Lieleg, O. Selected metal ions protect Bacillus subtilis biofilms from erosion. Metallomics 6, 1441–1450 (2014).
Cao, B., Ahmed, B. & Beyenal, H. in Emerging Environmental Technologies Vol. 2 (ed Shah, V.) 1–37 (Springer, 2010).
Hammaini, A., Gonzalez, F., Ballester, A., Bláquez, M. L. & Muñoz, J. A. Biosorption of heavy metals by activated sludge and their desorption characteristics. J. Environ. Manage. 84, 419–426 (2007).
Henze, M., van Loosdrechte, M., Ekama, G. A. & Brdjanovic, D. (eds) Biological Wastewater Treatment: Principles, Modelling and Design (IWA Publishing, 2008).
Okabe, S., Yasuda, T. & Watanabe, Y. Uptake and release of inert fluorescent particles by mixed population biofilms. Biotechnol. Bioeng. 53, 459–469 (1997).
Kouzuma, A., Kato, S. & Watanabe, K. Microbial interspecies interactions: recent findings in syntrophic consortia. Front. Microbiol. 6, 477 (2015).
Ikuma, K., Decho, A. W. & Lau, B. L. When nanoparticles meet biofilms — interactions guiding the environmental fate and accumulation of nanoparticles. Front. Microbiol. 6, 591 (2015).
Tielen, P. et al. Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa. BMC Microbiol. 159, 221–228 (2013).
Zobell, C. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 46, 39–56 (1943).
Wingender, J. & Jaeger, K.-E. in Encyclopedia of Environmental Microbiology Vol. 3 (ed. Bitton, G.) 1207–1223 (Wiley-Interscience, 2002).
Burns, R. G. Enzyme activity in soil: location and possible role in microbial ecology. Soil Biol. Biochem. 15, 423–427 (1982).
Lock, M. A., Wallace, R. R., Costerton, J. W., Ventullo, R. M. & Charlton, S. E. River epilithon: toward a structural–functional model. Oikos 42, 10–22 (1984).
Toyofuku, M., Roschitzki, B., Riedel, K. & Eberl, L. Identification of proteins associated with the Pseudomonas aeruginosa biofilom extracellular matrix. J. Proteome Res. 11, 4906–4915 (2012).
Zhang, W. et al. Extracellular matrix-associated proteins form an integral and dynamic system during Pseudomonas aeruginosa biofilm development. Front. Cell. Infect. Microbiol. 5, 40 (2015).
Worm, J., Jensen, L. E., Hansen, T. S., Søndergaard, M. & Nybroe, O. Interactions between proteolytic and non-proteolytic Pseudomonas fluorescens affect protein degradation in a model community. FEMS Microbiol. Ecol. 32, 103–109 (2000).
Smucker, R. A. & Kim, C. K. in Microbial Enzymes in Aquatic Environments (ed Chróst, R. J.) 249–269 (Springer–Verlag,1991).
Chang, Y.-W. et al. Biofilm formation in geometries with different surface curvature and oxygen availability. New J. Phys. 17, 033017 (2015).
Kalmbach, S., Manz, W. & Szewzyk, U. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63, 4164–4170 (1997).
Boles, B. R., Thoendel, M. & Singh, P. K. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl Acad. Sci. USA 101, 16630–16635 (2004). This paper reports the important finding that, unlike their planktonic counterparts, individual cells in the biofilm develop clearly distinct phenotypes after 1–2 days, even for biofilms that are formed by monoclonal organisms.
Von Ohle, C. et al. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl. Environ. Microbiol. 76, 2326–2334 (2010).
Kragh, K. N. et al. Role of multicellular aggregates in biofilm formation. mBio 7, e00237-16 (2016).
Ward, D. M. et al. Genomics, environmental genomics and the issue of microbial species. Heredity 100, 207–219 (2008).
Neu, T. R. & Lawrence, J. R. Advanced techniques for in situ analysis of the biofilm matrix (structure, composition, dynamics) by means of laser scanning microscopy. Methods Mol. Biol. 1147, 43–64 (2015).
Parsek, M. R. & Greenberg, E. P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33 (2005).
West, S. A., Griffin, A. S., Gardner, A. & Diggle, S. P. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 (2006).
Fredrickson, J. K. Ecological communities by design. Science 348, 1425–1427 (2015).
Zelezniak, A. et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl Acad. Sci. USA 112, 6449–6454 (2015).
Elias, S. & Banin, E. Multi-species biofilms: living with friendly neighbours. FEMS Microbiol. Rev. 36, 990–1004 (2012).
Koch, H. et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl Acad. Sci. USA 112, 11371–11376 (2015).
Gorbushina, A. A. & Broughton, W. J. Microbiology of the atmosphere–rock interface: how biological reactions and physical stress modulate a sophisticated microbial ecosystem. Annu. Rev. Microbiol. 63, 431–450 (2009).
Lee, K. W. et al. Biofilm development and enhanced stress resistance of a model, mixed species community biofilm. ISME J. 8, 894–907 (2014).
Breugelmans, P. et al. Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol. Ecol. 64, 271–282 (2008).
Hansen, S. K., Rainey, P. B., Haagensen, J. A. & Molin, S. Evolution of species interactions in a biofilm community. Nature 445, 533–536 (2007).
Ren, D., Madsen, J. S., Sørensen, S. & Burmølle, M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 9, 81–89 (2015).
Burmølle, M., Ren, D., Bjarnsholt, T. & Soerensen, S. J. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 22, 84–90 (2014).
Foster, K. R. & Bell, T. Competition, not cooperation dominates interactions among microbial species. Curr. Biol. 19, 1845–1850 (2012).
Rendueles, O. & Ghigo, J.-M. Mechanisms of competition in biofilm communities. Microbiol. Spectr. 3, MB-0009-2014 (2015).
McIntyre, D. L., Miyata, S. T., Kitaoka, M. & Pukazki, S. The Vibrio cholera type VI secretion system displays antimicrobial properties. Proc. Natl Acad. Sci. USA 107, 19520–19524 (2010).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886 (2015).
Thuptimdang, P., Limpiyakorn, T., McEvoy, J., Prüß, B. M. & Khan, E. Effect of silver nanoparticles on Pseudomonas putida biofilms at different stages of maturity. J. Hazard. Mater. 290, 127–133 (2015).
Königs, A. M., Flemming, H.-C. & Wingender, J. Nanosilver induces a non-culturable but metabolically active state in Pseudomonas aeruginosa. Front. Microbiol. 6, 395 (2015).
Oubekka, S. D., Briandet, R., Fontaine-Aupart, M.-P. & Steenkeste, K. Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrob. Agents Chemother. 56, 3349–3358 (2012). A paper that studies, in detail, the interactions between antibiotics and EPS components.
Ordax, M., Marco-Noales, E., López, M. M. & Biscoa, E. G. Exopolysaccharides favor the survival of Erwinia amylovora under copper stress through different strategies. Res. Microbiol. 161, 549–555 (2010).
Harrison, J., Ceri, H. & Turner, R. J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5, 928–939 (2007).
Khan, W. et al. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 13, 207–212 (2010).
Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005).
Brown, M. R., Allison, D. G. & Gilbert, P. Resistance of bacterial biofilms: a growth-related effect? J. Antimicrob. Chemother. 22, 777–783 (1988).
Amato, S. M. et al. The role of metabolism in bacterial persistence. Front. Microbiol. 5, 70 (2014).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014).
Monzón, M., Oteiza, C., Leiva, J., Lamata, M. & Amorena, B. Biofilm testing of Staphylococcus epidermidis clinical isolates: low performance of vancomycin in relation to other antibiotics. Diagn. Microbiol. Infect. Dis. 44, 319–324 (2002).
Li, L., Mendis, N., Trigui, H., Oliver, J. D. & Faucher, S. P. The importance of the viable-but-nonculturable state in human bacterial pathogens. Front. Microbiol. 5, 258 (2014).
Conlon, B. P., Rowe, S. E. & Lewis, K. Persister cells in biofilm associated infections. Adv. Exp. Med. Biol. 831, 1–9 (2015).
Ayrapetyan, M., Williams, T. C. & Oliver, J. D. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13 (2015).
Helaine, S. & Kugelberg, E. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol. 22, 417–424 (2014).
Keren, I., Minami, S., Rubin, E. & Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2, e00100-11 (2011).
Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 7, 1061–1072 (2012).
Madsen, J. S., Burmølle, M., Hansen, H. L. & Sørensen, S. J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195 (2012).
van Meervenne, E. et al. Biofilm models for the food industry: hot spots for plasmid transfer? Pathog. Dis. 70, 332–338 (2014).
Król, J. E. et al. Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 70, 110–119 (2013).
Savage, V. J., Chopra, I. & O'Neill, A. J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 57, 1968–1970 (2013).
Borgeaud, S., Metzger, L. C., Scrignari, T. Blokesch, M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015).
Wang, B. Y., Chi, B. & Kuramitsu, H. K. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 17, 108–112 (2002).
Merod, R. T. & Wuertz, S. Extracellular polymeric substance architecture influences natural genetic transformation of Acinetobacter baylyi in biofilms. Appl. Environ. Microbiol. 80, 7752–7757 (2014). An important paper regarding the role of EPS in facilitating horizontal gene transfer.
Karatan, E. & Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 (2009).
Thomsen, M. S. et al. Habitat cascades: The conceptual context and global relevance of facilitation cascades via habitat formation and modification. Integr. Comp. Biol. 50, 158–175 (2010).
Jones, C. G., Lawton, J. H. & Shachak, M. Organisms as ecosystem engineers. Oikos 69, 373–386 (1994).
Bennett, S. R. et al. The 'Great Southern Reef': social, ecological and economic value of Australia's neglected kelp forests. Mar. Freshw. Res. 67, 47–56 (2015).
Steneck, R. S. et al. Kelp forest ecosystems: biodiversity, stability, resilience and future. Environ. Conserv. 29, 436–459 (2002).
Lawrence, D. & Vandecar, K. Effects of tropical deforestation on climate change and agriculture. Nat. Clim. Chang. 5, 27–36 (2015).
Bruno, J. F., Stachowicz, J. J. & Bertness, M. D. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125 (2003).
Cornwall, C. E. et al. Diffusion boundary layers ameliorate the negative effects of ocean acidification on the temperate coralline macroalga Arthrocardia corymbosa. PLoS ONE 9, e97235 (2014).
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D. & Kjelleberg, S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 (2012).
Whitham, T. G. et al. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523 (2006).
Ponge, J.-F. Emergent properties from organisms to ecosystems: towards a realistic approach. Biol. Rev. 80, 403–411 (2005).
Lowman, M. D. & Schowalter, T. D. Plant science in forest canopies — the first 30 years of advances and challenges (1980-2010). New Phytol. 194, 12–27 (2012).
Matz, C. & Kjelleberg, S. Off the hook — how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307 (2005).
Aikens, K. R., Timms, L. L. & Buddle, C. M. Vertical heterogeneity in predation pressure in a temperate forest canopy. PeerJ 1, e138 (2013).
Hubbell, S. P. et al. Light-gap disturbances, recruitment limitation, and tree diversity in a neotropical forest. Science 283, 554–557 (1999).
Nealson, K. H. Autoinduction of bacterial luciferase. Arch. Microbiol. 112, 73–79 (1977). A pioneering and seminal study of intercellular communication in bacteria.
Keller, L. & Surette, M. G. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258 (2006).
Seviour, T. et al. Functional amyloids keep quorum sensing molecules in check. J. Biol. Chem. 290, 6457–6469 (2015).
Redfield, R. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10, 365–370 (2002).
Hense, B. A. et al. Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239 (2007).
Charlton, T. S. et al. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ. Microbiol. 2, 530–541 (2000).
Tan, C. H. et al. Community quorum sensing signalling and quenching: microbial granular biofilm assembly. NPJ Biofilms Microbiomes 1, 15006 (2015).
Kim, M., Ingremeau, F., Zhao, A., Bassler, B. L. & Stone, H. A. Local and global consequences of flow on bacterial quorum sensing. Nat. Microbiol. 1, 15005 (2016). This study reports a very interesting correlation between the activation of quorum sensing and external flow, explaining the spatially and temporally non-uniform responses to quorum sensing signals that are observed in natural environments.
Liu, J. et al. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554 (2015).
Prindle, A. et al. Ion channels enable electrical communication in bacterial community. Nature 527, 59–65 (2015). The discovery of an exciting alternative signalling mechanism to chemical communication among bacteria in biofilms.
Kato, S., Hashimoto, K. & Watanabe, K. Microbial interspecies electron transfer via electric currents through conductive materials. Proc. Natl Acad. Sci. USA 109, 10042–10046 (2012).
Balsalobre, C. et al. Release of the type I secreted α-haemolysin via outer membrane vesicles from Escherichia coli. Mol. Microbiol. 59, 99–112 (2006).
Acknowledgements
H.-C.F., S.K., S.R. and P.S. gratefully acknowledge the influence of the inspiring and motivating atmosphere during the Singapore Centre on Environmental Life Sciences Engineering (SCELSE) Summer Course while preparing this Review article. The authors thank J. Froesler for assistance with the figures. H.-C.F. appreciates the long-term support of the IWW Water Centre (Muelheim, Germany) to his work, and the support of P. Wilderer, his first mentor. The authors also thank SCELSE for supporting this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Grazing
-
A form of predation, such as when protozoa feed on bacteria.
- Macrophytes
-
Aquatic photosynthetic organisms that are visible to the naked eye. In marine systems, 'macrophytes' is often used as a general term that includes macroalgae, such as kelps, and coastal plants, such as seagrasses or mangroves.
- Bromeliads
-
A family of flowering plants that has leaves that often form in such a way so as to enable the persistence of pools of water that form distinct habitats and ecological communities.
- Microrheology
-
The study of the rheological properties of a material at the micrometre scale.
- Desiccation tolerance
-
The ability to survive water limitation.
- Nanowires
-
Electrically conductive structures that are produced by microorganisms.
- Rheologocial
-
Pertaining to the study of the flow of matter, primarily in a liquid state but also as 'soft solids' or solids under conditions in which they respond to an applied force with plastic flow rather than elastic deformation.
- Ecomechanics
-
The biomechanical mechanisms by which organisms interact with their environment.
- Electrogenic
-
Capable of generating an electric current.
- Microbial mats
-
A coherent, layered organization of microorganisms with complementary metabolic capacities. Microbial mats are typically found in aquatic environments, anchored to a surface.
- Mass transfer
-
The net movement of compounds or chemical species from one position to another.
- Streamers
-
Portions of the biofilm extracellular polymeric substances (EPS) that extend out from the biofilm surface into the liquid flow.
- Sorption
-
Adsorption or absorption, or a combination of both processes.
- Oligotrophic
-
An environment that is characterized by low nutrient concentrations.
- Lithification
-
A process in which sediments compact under pressure and gradually become solid rock. The biogenesis of carbonate can support this process.
- Activated sludge
-
The microbial biomass in the aerobic portion of a wastewater treatment system.
- Carbon cloth
-
A soft, flexible cloth-like material made from carbon fibre.
- Planktonic cells
-
Free-living cells (that is, not in a biofilm) in the liquid phase.
- Humic substances
-
The combination of compounds, generally humic acids, that make up the organic components of soils, peat and coal.
- Molecular modelling
-
Methods that encompass theoretical methods and computational techniques that are used to model or mimic the behaviour of molecules.
- Copiotrophic
-
An environment that is characterized by high nutrient concentrations.
- Heterotrophic
-
Refers to the use of organic compounds for nutrition.
- Type VI secretion systems
-
Multiprotein complexes that use a one-step mechanism to inject effector proteins, such as virulence factors, from the interior of a bacterial cell into a target cell. These systems have been found in a quarter of all proteobacterial genomes, including those that encode animal, plant and human pathogens, as well as soil, environmental and marine bacteria.
- Aminoglycosides
-
A class of antibiotics that typically have a cyclohexane ring and amino sugars.
- Stationary phase
-
A slow or non-growth phase of the bacterial growth cycle that typically results from a lack of electron donors or acceptors.
- Viable-but-nonculturable state
-
(VBNC state). A state of dormancy that is defined by failure of an organism to be cultured on media that normally supports its growth, while retaining measurable indications of viability, such as respiratory activity, the presence of rRNA and integrity of the cell membrane.
- Plasmid conjugation
-
The transfer of a specialized type of plasmid from one cell to another by a pilus that is encoded within the plasmid genome.
Rights and permissions
About this article
Cite this article
Flemming, HC., Wingender, J., Szewzyk, U. et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14, 563–575 (2016). https://doi.org/10.1038/nrmicro.2016.94
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.94
This article is cited by
-
Combination antimicrobial therapy: in vitro synergistic effect of anti-staphylococcal drug oxacillin with antimicrobial peptide nisin against Staphylococcus epidermidis clinical isolates and Staphylococcus aureus biofilms
Annals of Clinical Microbiology and Antimicrobials (2024)
-
In vitro antibacterial and anti-biofilm potential of an endophytic Schizophyllum commune
AMB Express (2024)
-
Anti-biofilm properties of laser-synthesized, ultrapure silver–gold-alloy nanoparticles against Staphylococcus aureus
Scientific Reports (2024)
-
Volatile methyl jasmonate from roots triggers host-beneficial soil microbiome biofilms
Nature Chemical Biology (2024)
-
Biofilm volume and acidification within initial biofilms formed in situ on buccally and palatally exposed bracket material
Journal of Orofacial Orthopedics / Fortschritte der Kieferorthopädie (2024)