Key Points
-
Poly(ADP-ribose) polymerase 1 (PARP1) was the first member of the PARP family to be identified. The PARP family now comprises 18 members.
-
PARP1 post-translationally modifies itself and a range of other proteins that have diverse roles in different cellular processes.
-
The catalytic activity of PARP1 is responsible for mediating multiple DNA damage repair pathways.
-
PARP1 has a crucial role in the stabilization of DNA replication forks.
-
The role of PARP1 in remodelling chromatin overlaps with its role in DNA repair.
-
PARP1 inhibition is an attractive strategy for the treatment of cancers that are deficient in the repair of DNA double-strand breaks by homologous recombination.
Abstract
Cells are exposed to various endogenous and exogenous insults that induce DNA damage, which, if unrepaired, impairs genome integrity and leads to the development of various diseases, including cancer. Recent evidence has implicated poly(ADP-ribose) polymerase 1 (PARP1) in various DNA repair pathways and in the maintenance of genomic stability. The inhibition of PARP1 is therefore being exploited clinically for the treatment of various cancers, which include DNA repair-deficient ovarian, breast and prostate cancers. Understanding the role of PARP1 in maintaining genome integrity is not only important for the design of novel chemotherapeutic agents, but is also crucial for gaining insights into the mechanisms of chemoresistance in cancer cells. In this Review, we discuss the roles of PARP1 in mediating various aspects of DNA metabolism, such as single-strand break repair, nucleotide excision repair, double-strand break repair and the stabilization of replication forks, and in modulating chromatin structure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tubbs, A. & Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017).
Lord, C. J., Tutt, A. N. & Ashworth, A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 66, 455–470 (2015).
Montoni, A., Robu, M., Pouliot, E. & Shah, G. M. Resistance to PARP-inhibitors in cancer therapy. Front. Pharmacol. 4, 18 (2013).
Ame, J. C., Spenlehauer, C. & de Murcia, G. The PARP superfamily. Bioessays 26, 882–893 (2004).
Buki, K. G. & Kun, E. Polypeptide domains of ADP-ribosyltransferase obtained by digestion with plasmin. Biochemistry 27, 5990–5995 (1988).
Froelich, C. J. et al. Granzyme B/perforin-mediated apoptosis of Jurkat cells results in cleavage of poly(ADP-ribose) polymerase to the 89-kDa apoptotic fragment and less abundant 64-kDa fragment. Biochem. Biophys. Res. Commun. 227, 658–665 (1996).
Kameshita, I., Matsuda, Z., Taniguchi, T. & Shizuta, Y. Poly (ADP-ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J. Biol. Chem. 259, 4770–4776 (1984).
Langelier, M. F., Planck, J. L., Roy, S. & Pascal, J. M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012). This article reports the crystal structure of PARP1 bound to a DNA DSB and proposes a mechanism for the DNA-dependent activation of PARP1.
Nishikimi, M., Ogasawara, K., Kameshita, I., Taniguchi, T. & Shizuta, Y. Poly(ADP-ribose) synthetase. The DNA binding domain and the automodification domain. J. Biol. Chem. 257, 6102–6105 (1982).
Bork, P. et al. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11, 68–76 (1997).
D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999).
Kraus, W. L. & Lis, J. T. PARP goes transcription. Cell 113, 677–683 (2003).
Kim, M. Y., Zhang, T. & Kraus, W. L. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 (2005).
Hanzlikova, H., Gittens, W., Krejcikova, K., Zeng, Z. & Caldecott, K. W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 45, 2546–2557 (2017).
Isabelle, M. et al. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 8, 22 (2010).
Menissier de Murcia, J. et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 (2003).
Huambachano, O., Herrera, F., Rancourt, A. & Satoh, M. S. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 286, 7149–7160 (2011).
Gagne, J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976 (2008).
Jungmichel, S. et al. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 52, 272–285 (2013). This article reports the high-throughput identification of targets of PARylation in response to different genotoxic stresses.
Hassa, P. O. & Hottiger, M. O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 (2008).
Krietsch, J. et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 34, 1066–1087 (2013).
Teloni, F. & Altmeyer, M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 44, 993–1006 (2016).
Althaus, F. R. et al. Poly ADP-ribosylation: a DNA break signal mechanism. Mol. Cell. Biochem. 193, 5–11 (1999).
Malanga, M., Pleschke, J. M., Kleczkowska, H. E. & Althaus, F. R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 273, 11839–11843 (1998).
Pleschke, J. M., Kleczkowska, H. E., Strohm, M. & Althaus, F. R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000).
Meyer-Ficca, M. L., Meyer, R. G., Coyle, D. L., Jacobson, E. L. & Jacobson, M. K. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297, 521–532 (2004).
Erdelyi, K. et al. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 23, 3553–3563 (2009).
Feng, X. & Koh, D. W. Inhibition of poly(ADP-ribose) polymerase-1 or poly(ADPribose) glycohydrolase individually, but not in combination, leads to improved chemotherapeutic efficacy in HeLa cells. Int. J. Oncol. 42, 749–756 (2013).
Fisher, A. E. O., Hochegger, H., Takeda, S. & Caldecott, K. W. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 27, 5597–5605 (2007).
Koh, D. W. et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA 101, 17699–17704 (2004). This article reports the essential role of PARG in degradation of PAR.
Ray Chaudhuri, A., Ahuja, A. K., Herrador, R. & Lopes, M. Poly(ADP-ribosyl) glycohydrolase prevents the accumulation of unusual replication structures during unperturbed S phase. Mol. Cell. Biol. 35, 856–865 (2015).
Zhou, Y., Feng, X. & Koh, D. W. Enhanced DNA accessibility and increased DNA damage induced by the absence of poly(ADP-ribose) hydrolysis. Biochemistry 49, 7360–7366 (2010).
Caldecott, K. W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 (2008).
Satoh, M. S. & Lindahl, T. Role of poly(Adp-ribose) formation in DNA-repair. Nature 356, 356–358 (1992).
Caldecott, K. W., McKeown, C. K., Tucker, J. D., Ljungquist, S. & Thompson, L. H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14, 68–76 (1994).
Loizou, J. I. et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117, 17–28 (2004).
Marintchev, A. et al. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 28, 2049–2059 (2000).
Whitehouse, C. J. et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104, 107–117 (2001).
El-Khamisy, S. F., Masutani, M., Suzuki, H. & Caldecott, K. W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31, 5526–5533 (2003). This article shows the requirement for PARP1 in the recruitment of XRCC1, which is an essential factor in the repair of SSBs.
Schreiber, V. et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277, 23028–23036 (2002).
Hoch, N. C. et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91 (2017).
Oei, S. L. & Ziegler, M. ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose). J. Biol. Chem. 275, 23234–23239 (2000).
Petermann, E., Ziegler, M. & Oei, S. L. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair 2, 1101–1114 (2003).
Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Pouliot, J. J., Yao, K. C., Robertson, C. A. & Nash, H. A. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555 (1999).
Yang, S. W. et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA 93, 11534–11539 (1996).
Das, B. B. et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 42, 4435–4449 (2014). This article elucidates the interaction of PARP1 and TDP1 and its important role in the repair of TOP1-abortive complexes.
Patel, A. G. et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J. Biol. Chem. 287, 4198–4210 (2012).
Dantzer, F. et al. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39, 7559–7569 (2000).
Dantzer, F. et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81, 69–75 (1999).
de Murcia, J. M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA 94, 7303–7307 (1997).
Pachkowski, B. F. et al. Cells deficient in PARP-1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP-1-proficient cells exposed to MMS. Mutat. Res. 671, 93–99 (2009).
Vodenicharov, M. D., Sallmann, F. R., Satoh, M. S. & Poirier, G. G. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 28, 3887–3896 (2000).
Wang, Z. Q. et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11, 2347–2358 (1997).
Allinson, S. L., Dianova, I. I. & Dianov, G. L. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol. 50, 169–179 (2003).
Orta, M. L. et al. The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic Acids Res. 42, 9108–9120 (2014).
Strom, C. E. et al. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 39, 3166–3175 (2011).
Reynolds, P., Cooper, S., Lomax, M. & O'Neill, P. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 43, 4028–4038 (2015).
Marteijn, J. A., Lans, H., Vermeulen, W. & Hoeijmakers, J. H. J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014).
Guerrero-Santoro, J. et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68, 5014–5022 (2008).
Kapetanaki, M. G. et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl Acad. Sci. USA 103, 2588–2593 (2006).
Wang, H. et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 (2006).
Pines, A. et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 199, 235–249 (2012). This report shows the role of PARP1 in mediating NER through the recruitment of DDB2 and the chromatin modifier ALC1.
Robu, M. et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl Acad. Sci. USA 110, 1658–1663 (2013).
Luijsterburg, M. S. et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 197, 267–281 (2012).
King, B. S., Cooper, K. L., Liu, K. J. & Hudson, L. G. Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J. Biol. Chem. 287, 39824–39833 (2012).
Mehta, A. & Haber, J. E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 6, a016428 (2014).
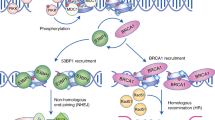
Chapman, J. R., Taylor, M. R. G. & Boulton, S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012).
Price, B. D. & D'Andrea, A. D. Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 (2013).
Ali, A. A. et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 19, 685–692 (2012).
Langelier, M. F. & Pascal, J. M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 23, 134–143 (2013).
Polo, S. E. & Jackson, S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011).
Sukhanova, M. V. et al. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 44, e60 (2016).
Haince, J. F. et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 282, 16441–16453 (2007).
Aguilar-Quesada, R. et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 8, 29 (2007).
Menisser-de Murcia, J., Mark, M., Wendling, O., Wynshaw-Boris, A. & de Murcia, G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol. 21, 1828–1832 (2001).
Haince, J. F. et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008).
Hochegger, H. et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25, 1305–1314 (2006).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Cruz-Garcia, A., Lopez-Saavedra, A. & Huertas, P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 9, 451–459 (2014).
Scully, R. et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997).
Li, M. & Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013).
Schwertman, P., Bekker-Jensen, S. & Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016).
Morgan, W. F. & Cleaver, J. E. 3-Aminobenzamide synergistically increases sister-chromatid exchanges in cells exposed to methyl methanesulfonate but not to ultraviolet light. Mutat. Res. 104, 361–366 (1982).
Oikawa, A., Tohda, H., Kanai, M., Miwa, M. & Sugimura, T. Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem. Biophys. Res. Commun. 97, 1311–1316 (1980).
Schultz, N., Lopez, E., Saleh-Gohari, N. & Helleday, T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 31, 4959–4964 (2003).
Yang, Y. G., Cortes, U., Patnaik, S., Jasin, M. & Wang, Z. Q. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882 (2004).
Hu, Y. et al. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 4, 1430–1447 (2014).
El-Khamisy, S. F. et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005).
Fan, J. et al. XRCC1 down-regulation in human cells leads to DNA-damaging agent hypersensitivity, elevated sister chromatid exchange, and reduced survival of BRCA2 mutant cells. Environ. Mol. Mutagen. 48, 491–500 (2007).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 (2010).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). References 92 and 93 are the first reports of the synthetic lethality of the combined loss of PARP1 and BRCA2.
Evers, B., Helleday, T. & Jonkers, J. Targeting homologous recombination repair defects in cancer. Trends Pharmacol. Sci. 31, 372–380 (2010).
Rottenberg, S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17079–17084 (2008).
Ruscetti, T. et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 273, 14461–14467 (1998).
Spagnolo, L., Barbeau, J., Curtin, N. J., Morris, E. P. & Pearl, L. H. Visualization of a DNA-PK/PARP1 complex. Nucleic Acids Res. 40, 4168–4177 (2012).
Rybanska, I. et al. PARP1 and DNA-PKcs synergize to suppress p53 mutation and telomere fusions during T-lineage lymphomagenesis. Oncogene 32, 1761–1771 (2013).
Luijsterburg, M. S. et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 (2016).
Truong, L. N. et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. USA 110, 7720–7725 (2013).
Deriano, L. & Roth, D. B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455 (2013).
Yan, C. T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007).
Cheng, Q. et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res. 39, 9605–9619 (2011).
Fattah, F. et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 6, e1000855 (2010).
Mansour, W. Y., Rhein, T. & Dahm-Daphi, J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 38, 6065–6077 (2010).
Wang, M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006).
Wray, J. et al. PARP1 is required for chromosomal translocations. Blood 121, 4359–4365 (2013).
Celli, G. B. & de Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 (2005).
Celli, G. B., Denchi, E. L. & de Lange, T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8, 885–890 (2006).
Sfeir, A. & de Lange, T. Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597 (2012).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518, 258–262 (2015).
Kent, T., Mateos-Gomez, P. A., Sfeir, A. & Pomerantz, R. T. Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, e13740 (2016).
Mateos-Gomez, P. A. et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). References 111 and 113 discuss the requirement for Pol θ in aNHEJ.
Anachkova, B., Russev, G. & Poirier, G. G. DNA replication and poly(ADP-ribosyl)ation of chromatin. Cytobios 58, 19–28 (1989).
Lehmann, A. R., Kirk-Bell, S., Shall, S. & Whish, W. J. The relationship between cell growth, macromolecular synthesis and poly ADP-ribose polymerase in lymphoid cells. Exp. Cell Res. 83, 63–72 (1974).
Bryant, H. E. et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28, 2601–2615 (2009).
Dantzer, F., Nasheuer, H. P., Vonesch, J. L., de Murcia, G. & Menissier-de Murcia, J. Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 26, 1891–1898 (1998).
Simbulan-Rosenthal, C. M. et al. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry 37, 9363–9370 (1998).
Smirnova, M. & Klein, H. L. Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat. Res. 532, 117–135 (2003).
Dungrawala, H. et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015).
Ray Chaudhuri, A. et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19, 417–423 (2012). This report demonstrates the essential role of PARP1 in replication fork reversal.
Sugimura, K., Takebayashi, S., Taguchi, H., Takeda, S. & Okumura, K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 183, 1203–1212 (2008).
Zellweger, R. et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208, 563–579 (2015).
Follonier, C., Oehler, J., Herrador, R. & Lopes, M. Friedreich's ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat. Struct. Mol. Biol. 20, 486–494 (2013).
Neelsen, K. J. & Lopes, M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16, 207–220 (2015).
Neelsen, K. J., Zanini, I. M., Herrador, R. & Lopes, M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200, 699–708 (2013).
Atkinson, J. & McGlynn, P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37, 3475–3492 (2009).
Berti, M. et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20, 347–354 (2013).
Ahuja, A. K. et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7, 10660 (2016).
Ding, X. et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 7, 12425 (2016). This report shows that synthetic viability results from deficiency of both PARP1 and BRCA2.
Ying, S., Hamdy, F. C. & Helleday, T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72, 2814–2821 (2012).
Schlacher, K. et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012).
Petermann, E., Orta, M. L., Issaeva, N., Schultz, N. & Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010).
Gottipati, P. et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 70, 5389–5398 (2010).
Ray Chaudhuri, A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016). This report demonstrates that a loss of PARP1 that precedes BRCA1 loss results in relative genome stability owing to replication fork protection.
Illuzzi, G. et al. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res. 42, 7776–7792 (2014).
Guillemette, S. et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 29, 489–494 (2015).
Escargueil, A. E., Soares, D. G., Salvador, M., Larsen, A. K. & Henriques, J. A. What histone code for DNA repair? Mutat. Res. 658, 259–270 (2008).
Gibson, B. A. & Kraus, W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012).
Kraus, W. L. & Hottiger, M. O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 34, 1109–1123 (2013).
Schiewer, M. J. & Knudsen, K. E. Transcriptional roles of PARP1 in cancer. Mol. Cancer Res. 12, 1069–1080 (2014).
Messner, S. et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38, 6350–6362 (2010).
Poirier, G. G., de Murcia, G., Jongstra-Bilen, J., Niedergang, C. & Mandel, P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982).
Chen, M. et al. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS ONE 4, e6727 (2009).
Ma, N. F. et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 47, 503–510 (2008).
Ahel, D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009). This report shows that PARP1-dependent chromatin remodelling by ALC1 is essential for DNA repair.
Gottschalk, A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009).
Smeenk, G. et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 126, 889–903 (2013).
Malewicz, M. et al. Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 25, 2031–2040 (2011).
Kruhlak, M. et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734 (2007).
Shanbhag, N. M., Rafalska-Metcalf, I. U., Balane-Bolivar, C., Janicki, S. M. & Greenberg, R. A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010).
Chou, D. M. et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA 107, 18475–18480 (2010).
Polo, S. E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S. P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010).
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015).
Vinayak, S. & Ford, J. M. PARP inhibitors for the treatment and prevention of breast cancer. Curr. Breast Cancer Rep. 2, 190–197 (2010).
Gagne, J. P., Hendzel, M. J., Droit, A. & Poirier, G. G. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr. Opin. Cell Biol. 18, 145–151 (2006).
Min, W. & Wang, Z. Q. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front. Biosci. (Landmark Ed.) 14, 1619–1626 (2009).
Barkauskaite, E., Jankevicius, G. & Ahel, I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell 58, 935–946 (2015).
Andrabi, S. A. et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA 103, 18308–18313 (2006).
Mortusewicz, O., Fouquerel, E., Ame, J. C., Leonhardt, H. & Schreiber, V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 39, 5045–5056 (2011).
Cortes, U. et al. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol. 24, 7163–7178 (2004).
Mueller-Dieckmann, C. et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006).
Oka, S., Kato, J. & Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 (2006).
Rosenthal, F. et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 20, 502–507 (2013).
Sharifi, R. et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 (2013).
Krishnakumar, R. et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 (2008).
Hassa, P. O., Buerki, C., Lombardi, C., Imhof, R. & Hottiger, M. O. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 278, 45145–45153 (2003).
Gibson, B. A. et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50 (2016).
Soldatenkov, V. A. et al. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J. Biol. Chem. 277, 665–670 (2002).
Acknowledgements
The authors are especially grateful to A. Tubbs, S. John and G. Poirier for comments on the manuscript and also for discussions. This work was supported by the Intramural Research Program of the US National Institutes of Health (NIH), the US National Cancer Institute and the Center for Cancer Research. A.N. was also supported by the US Department of Defense (BCRP DOD Idea Expansion Award BC133858 and BCRP Breakthrough Award BC151331), the Ellison Foundation Award for Aging Research and Alex's Lemonade Stand Foundation Reach Award. A.R.C. has been supported by a Human Frontier Science Program Long-term Fellowship (LT000393/2013).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
Glossary
- DNA damage response
-
(DDR). The collection of cellular pathways that detect, signal and repair DNA damage.
- BRCT domain
-
(BRCA1 C terminus domain). An evolutionarily conserved protein domain that has DNA repair functions; it contains phosphoprotein-binding sites.
- Abasic sites
-
DNA sites that lack either a purine or a pyrimidine base owing to endogenous and/or exogenous DNA damage.
- Epistasis
-
A genetic interaction in which a mutation in one gene masks the effects of a mutation in another gene.
- Oxidative base damage
-
Damage to DNA bases caused by oxidation, which mostly modifies guanine to produce 8-hydroxyguanine.
- Alkylation damage
-
DNA damage mediated by transfer of a single methyl group to a DNA base (mostly to N or O atoms of guanine), which results in the formation of a methyl adduct on the base.
- Lesion verification
-
Verification of a chemical modification on the DNA by the transcription and repair factor transcription factor IIH (TFIIH) during nucleotide excision repair.
- Class-switch recombination
-
(CSR). A process in B cells that involves switching the type of antibody that is produced by changing the constant region of the antibody heavy chain.
- V(D)J recombination
-
A DNA recombination process that occurs during B cell or T cell activation, in which the variable domain exons of antigen receptors are assembled from sub-exonic segments called V, D and J to ultimately generate an immunoglobulin gene or T cell receptor, respectively.
- One-ended DSBs
-
(One-ended DNA double-strand breaks). DSBs formed during collision of ongoing replications forks with a lesion on one strand of the template DNA.
- Shelterin complex
-
A complex of six proteins that binds to TTAGGG repeats at telomeres and protects them from recognition as DNA double-strand breaks.
- Terminal transferase
-
An enzyme that catalyses the addition of nucleotides to 3′ DNA overhangs at double-stranded DNA.
Rights and permissions
About this article
Cite this article
Ray Chaudhuri, A., Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 18, 610–621 (2017). https://doi.org/10.1038/nrm.2017.53
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.53
This article is cited by
-
Co-Packaged PARP inhibitor and photosensitizer for targeted photo-chemotherapy of 3D ovarian cancer spheroids
Cell & Bioscience (2024)
-
Physiology and pharmacological targeting of phase separation
Journal of Biomedical Science (2024)
-
T-type voltage-gated channels, Na+/Ca2+-exchanger, and calpain-2 promote photoreceptor cell death in inherited retinal degeneration
Cell Communication and Signaling (2024)
-
Transcription–replication conflicts underlie sensitivity to PARP inhibitors
Nature (2024)
-
PARP1-stabilised FOXQ1 promotes ovarian cancer progression by activating the LAMB3/WNT/β-catenin signalling pathway
Oncogene (2024)