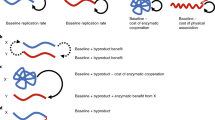
Key Points
-
It is often assumed that molecular systems are optimized to maximize the competitive ability of the organism.
-
Sociobiology predicts that molecular systems are shaped by cooperative and competitive phenotypes across multiple levels of biological organization.
-
Low-level selection for competition can disrupt social systems, as illustrated by transposable elements, cancer and disease.
-
High-level selection for cooperation can promote social systems, as illustrated by genomes, multicellularity and animal societies.
-
Cooperative and competitive forces shape four features of molecular systems: their functional scale, connections, diversity and rate of change.
-
The functional scale of molecular systems — the number of genes that work together towards a common goal — is shaped by the potential for competition.
-
The number and nature of connections in molecular networks is shaped by competition through links to modularity, robustness and the specificity of interactions.
-
Natural selection to generate diverse interacting phenotypes has strongly shaped the molecular networks of cooperation systems, including multicellular development.
-
Social evolution influences the rates at which molecular networks change in real and evolutionary time.
-
There is a strong case for integrating the theories of sociobiology into the study of molecular systems.
Abstract
It is often assumed that molecular systems are designed to maximize the competitive ability of the organism that carries them. In reality, natural selection acts on both cooperative and competitive phenotypes, across multiple scales of biological organization. Here I ask how the potential for social effects in evolution has influenced molecular systems. I discuss a range of phenotypes, from the selfish genetic elements that disrupt genomes, through metabolism, multicellularity and cancer, to behaviour and the organization of animal societies. I argue that the balance between cooperative and competitive evolution has shaped both form and function at the molecular scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Huxley, J. Evolution: the Modern Synthesis (Harper & Brothers, 1942).
Hamilton, W. D. The genetical evolution of social behaviour. I & II. J. Theor. Biol. 7, 1–52 (1964). This seminal paper shows how natural selection can favour the evolution of cooperative and altruistic phenotypes.
Wilson, D. S. A theory of group selection. Proc. Natl Acad. Sci. USA 72, 143–146 (1975).
Okasha, S. Evolution and the Levels of Selection (Oxford Univ. Press, USA, 2006). This book works through the theoretical basis for natural selection at multiple levels of biological organization.
Robinson, G. E. Integrative animal behaviour and sociogenomics. Trends Ecol. Evol. 14, 202–205 (1999).
Smith, C. R., Toth, A. L., Suarez, A. V. & Robinson, G. E. Genetic and genomic analyses of the division of labour in insect societies. Nature Rev. Genet. 9, 735–748 (2008).
Robinson, G. E., Grozinger, C. M. & Whitfield, C. W. Sociogenomics: social life in molecular terms. Nature Rev. Genet. 6, 257–270 (2005).
Owens, I. P. F. Where is behavioural ecology going? Trends Ecol. Evol. 21, 356–361 (2006).
Foster, K. R., Parkinson, K. & Thompson, C. R. What can microbial genetics teach sociobiology? Trends Genet. 23, 74–80 (2007).
Grafen, A. in Behavioural Ecology: An Evolutionary Approach (eds Krebs, J. R. & Davies, N. B.) 62–84 (Blackwell, Oxford, 1984).
Banga, J. Optimization in computational systems biology. BMC Syst. Biol. 2, 47 (2008).
Hartwell, L., Hopfield, J., Leibler, S. & Murray, A. From molecular to modular cell biology. Nature 402, C47–C52 (1999).
Gardner, A. Adaptation as organism design. Biol. Lett. 5, 861–864 (2009). This article discusses the links between adaptation and the appearance of design in biological systems.
Medina, M. Genomes, phylogeny, and evolutionary systems biology. Proc. Natl Acad. Sci. USA 102, 6630–6635 (2005).
Soyer, O. The promise of evolutionary systems biology: Lessons from bacterial chemotaxis. Sci. STKE 3, pe23 (2010).
Lynch, M. The evolution of genetic networks by non-adaptive processes. Nature Rev. Genet. 8, 803–813 (2007).
Grafen, A. A first formal link between the price equation and an optimization program. J. Theor. Biol. 217, 75–91 (2002).
Sutherland, W. The best solution. Nature 435, 569 (2005).
Krebs, J., Kacelnik, A. & Taylor, P. Test of optimal sampling by foraging great tits. Nature 275, 27–31 (1978).
Pérez-Escudero, A., Rivera-Alba, M. & de Polavieja, G. Structure of deviations from optimality in biological systems. Proc. Natl Acad. Sci. USA 106, 20544–20549 (2009).
Grafen, A. Optimization of inclusive fitness. J. Theor. Biol. 238, 541–563 (2006).
Gardner, A. & Grafen, A. Capturing the superorganism: a formal theory of group adaptation. J. Evol. Biol. 22, 659–671 (2009).
Foster, K. R. A defense of sociobiology. Cold Spring Harb. Symp. Quant. Biol. LXXIV, 403–418 (2009). A review of the debates in social evolution that highlights the common principles underlying all sociobiological theory.
El-Samad, H., Kurata, H., Doyle, J., Gross, C. & Khammash, M. Surviving heat shock: control strategies for robustness and performance. Proc. Natl Acad. Sci. USA 102, 2736–2741 (2005).
Wagner, G. P., Pavlicev, M. & Cheverud, J. M. The road to modularity. Nature Rev. Genet. 8, 921–931 (2007).
Burt, A. & Trivers, R. L. Genes in Conflict: The Biology of Selfish Genetic Elements (Harvard Univ. Press, Harvard, 2006).
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Proulx, S. R., Promislow, D. E. L. & Phillips, P. C. Network thinking in ecology and evolution. Trends Ecol. Evol. 20, 345–353 (2005).
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
Foster, K. R. & Wenseleers, T. A general model for the evolution of mutualisms. J. Evol. Biol. 19, 1283–1293 (2006).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Nadell, C. D., Xavier, J. B. & Foster, K. R. The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224 (2009).
Edwards, J. & Palsson, B. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics 1, 1 (2000).
Edwards, J., Ibarra, R. & Palsson, B. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotech. 19, 125–130 (2001).
Reed, T. et al. In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst. Biol. 3, 114 (2009).
Schuetz, R., Kuepfer, L. & Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3, 119 (2007). This paper on flux balance analysis shows that bacterial metabolism seems to be designed to solve more than one biological problem.
Postma, E., Verduyn, C., Scheffers, W. & Van Dijken, J. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55, 468–477 (1989).
Pfeiffer, T., Schuster, S. & Bonhoeffer, S. Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001). Theoretical study that shows the importance of social evolution for metabolic systems.
MacLean, R. C. & Gudelj, I. Resource competition and social conflict in experimental populations of yeast. Nature 441, 498–501 (2006).
Hsu, P. P. & Sabatini, D. M. Cancer cellmetabolism: Warburg and beyond. Cell 134, 703–707 (2008).
Warburg, O. Uber die letzte Ursache und die entfernten Ursachen des Krebses (Verlag K. Triltsch, Wurzburg, 1966); English translation in Burk, D. The Prime Cause and Prevention of Cancer (1969).
Tyler, A., Asselbergs, F., Williams, S. & Moore, J. Shadows of complexity: what biological networks reveal about epistasis and pleiotropy. BioEssays 31, 220–227 (2009).
Wagner, G. Homologues, natural kinds and the evolution of modularity. Integr. Comp. Biol. 36, 36–43 (1996).
Kashtan, N. & Alon, U. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA 102, 13773–13778 (2005).
Solé, R. & Valverde, S. Spontaneous emergence of modularity in cellular networks. J. R. Soc. Interface 5, 129–133 (2008).
Rumbaugh, K. P. et al. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19, 341–345 (2009).
Foster, K. R., Shaulsky, G., Strassmann, J. E., Queller, D. C. & Thompson, C. R. L. Pleiotropy as a mechanism to stabilize cooperation. Nature 431, 693–696 (2004).
Merlo, L. M., Pepper, J. W., Reid, B. J. & Maley, C. C. Cancer as an evolutionary and ecological process. Nature Rev. Cancer 6, 924–935 (2006).
Park, S. Y., Gonen, M., Kim, H. J., Michor, F. & Polyak, K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 120, 636–644 (2010).
Vogelstein, B. & Kinzler, K. Cancer genes and the pathways they control. Nature Med. 10, 789–799 (2004).
Reuschenbach, M., von Knebel Doeberitz, M. & Wentzensen, N. A systematic review of humoral immune responses against tumor antigens. Cancer Immunol. Immunother. 58, 1535–1544 (2009).
Krakauer, D. & Plotkin, J. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl Acad. Sci. USA 99, 1405–1409 (2002).
Wilke, C., Wang, J., Ofria, C., Lenski, R. & Adami, C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412, 331–333 (2001).
Salathe, M. & Soyer, O. S. Parasites lead to evolution of robustness against gene loss in host signaling networks. Mol. Syst. Biol. 4, 202 (2008). Theoretical study showing a link between parasites and the evolution of mutational robustness in hosts.
Wagner, A. & Wright, J. Alternative routes and mutational robustness in complex regulatory networks. Biosystems 88, 163–172 (2007).
Soyer, O. & Pfeiffer, T. Evolution under fluctuating environments explains observed robustness in metabolic networks. PLoS Comput. Biol. 6, e1000907 (2010).
Wagner, A. Distributed robustness versus redundancy as causes of mutational robustness. BioEssays 27, 176–188 (2005).
Ihmels, J., Collins, S., Schuldiner, M., Krogan, N. & Weissman, J. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol. Syst. Biol. 3, 86 (2007).
Macia, J. & Solé, R. Distributed robustness in cellular networks: insights from synthetic evolved circuits. J. R. Soc. Interface 6, 393–400 (2009).
Sachs, J. L. & Bull, J. J. Experimental evolution of conflict mediation between genomes. Proc. Natl Acad. Sci. USA 102, 390–395 (2005).
Smukalla, S. et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 727–736 (2008).
Garneau, J. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Marraffini, L. & Sontheimer, E. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Gardner, A., West, S. A. & Buckling, A. Bacteriocins, spite and virulence. Proc. Biol. Sci. 271, 1529–1535 (2004).
Cascales, E. et al. Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 (2007).
Nakayama, K. et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38, 213–231 (2000).
Kohler, T., Donner, V. & van Delden, C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J. Bacteriol. 192, 1921 (2010).
Nicotra, M. L. et al. A hypervariable invertebrate allodeterminant. Curr. Biol. 19, 583–589 (2009).
De Tomaso, A. W. et al. Isolation and characterization of a protochordate histocompatibility locus. Nature 438, 454–459 (2005).
Hedrick, P. W. Evolutionary genetics of the major histocompatibility complex. Am. Nat. 143, 945–964 (1994).
Siddle, H. V. et al. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc. Natl Acad. Sci. USA 104, 16221–16226 (2007).
Rousset, F. & Roze, D. Constraints on the origin and maintenance of genetic kin recognition. Evolution 61, 2320–2330 (2007).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Mandeville, B. The Fable of the Bees: of Private Vices, Public Benefits (Oxford: Clarendon Press, 1732).
Scheffer, M. & van Nes, E. H. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc. Natl Acad. Sci. USA 103, 6230–6235 (2006).
Shoresh, N., Hegreness, M. & Kishony, R. Evolution exacerbates the paradox of the plankton. Proc. Natl Acad. Sci. USA 105, 12365–12369 (2008).
Queller, D. C. Relatedness and the fraternal major transitions. Phil. Trans. R. Soc. Lond. B 355, 1647–1655 (2000). A discussion paper that highlights the differences between coalitions of genes and species versus coalitions of cells and organisms of one species.
Süel, G., Garcia-Ojalvo, J., Liberman, L. & Elowitz, M. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440, 545–550 (2006).
Elango, N., Hunt, B. G., Goodisman, M. A. D. & Yi, S. V. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl Acad. Sci. USA 106, 11206–11211 (2009).
Behura, S. K. & Whitfield, C. W. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol. Biol. 19, 431–439 (2010).
Kucharski, R., Maleszka, J., Foret, S. & Maleszka, R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827–1830 (2008). An RNAi study showing the importance of DNA methylation for queen–worker determination in honeybees.
Aamodt, R. M. The caste- and age-specific expression signature of honeybee heat shock genes shows an alternative splicing-dependent regulation of Hsp90. Mech. Ageing Dev. 129, 632–637 (2008).
Calarco, J. A. et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138, 898–910 (2009).
Penny, D., Hoeppner, M. P., Poole, A. M. & Jeffares, D. C. An overview of the introns-first theory. J. Mol. Evol. 69, 527–540 (2009).
Agrawal, A., Eastman, Q. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998).
Bourke, A. F. G. & Franks, N. R. Social Evolution in Ants (Princeton Univ. Press, Princeton, New Jersey, 1995).
Ratnieks, F. L. W., Foster, K. R. & Wenseleers, T. Conflict resolution in insect societies. Annu. Rev. Entomol. 51, 581–608 (2006).
Axelrod, R. & Hamilton, W. D. The evolution of cooperation. Science 211, 1390–1396 (1981).
Perkins, T. J. & Swain, P. S. Strategies for cellular decision-making. Mol. Syst. Biol. 5, 326 (2009).
Fuqua, W. C., Winans, S. C. & Greenberg, E. P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994).
Keller, L. & Surette, M. G. Communication in bacteria: an ecological and evolutionary perspective. Nature Rev. Microbiol. 4, 249–258 (2006).
Miller, M. B. & Bassler, B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199 (2001).
Xavier, J., Kim, W. & Foster, K. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79, 166–179 (2011). An empirical study that shows a link between transcriptional regulation and the stabilization of cooperative phenotypes against the evolution of selfish phenotypes.
Ketting, R. F., Haverkamp, T. H. A., van Luenen, H. & Plasterk, R. H. A. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141 (1999).
Korb, J., Weil, T., Hoffmann, K., Foster, K. R. & Rehli, M. A gene necessary for reproductive suppression in termites. Science 324, 758 (2009).
Hunt, B. G. et al. Sociality is linked to rates of protein evolution in a highly social insect. Mol. Biol. Evol. 27, 497–500 (2010).
Buttery, N. J., Rozen, D. E., Wolf, J. B. & Thompson, C. R. L. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Curr. Biol. 19, 1373–1377 (2009).
Santorelli, L. A. et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature 451, 1107–1110 (2008).
Fiegna, F., Yu, Y. T., Kadam, S. V. & Velicer, G. J. Evolution of an obligate social cheater to a superior cooperator. Nature 441, 310–314 (2006).
Khare, A. et al. Cheater-resistance is not futile. Nature 461, 980–982 (2009).
Foster, K. R. Sociobiology: the Phoenix effect. Nature 441, 291–292 (2006).
Borghans, J., Beltman, J. & Boer, R. MHC polymorphism under host-pathogen coevolution. Immunogenetics 55, 732–739 (2004).
Petersen, L., Bollback, J. P., Dimmic, M., Hubisz, M. & Nielsen, R. Genes under positive selection in Escherichia coli. Genome Res. 17, 1336–1343 (2007).
Dorus, S., Wasbrough, E. R., Busby, J., Wilkins, E. C. & Karr, T. L. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol. Biol. Evol. 27, 1235–1246 (2010).
Tuemmler, B. & Cornelis, P. Pyoverdine receptor: a case of positive Darwinian selection in Pseudomonas aeruginosa. J. Bacteriol. 187, 3289–3292 (2005).
Drummond, D. A. & Wilke, C. O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 (2008). A theoretical and bioinformatics paper that shows the importance of gene expression level for understanding evolutionary rates.
Julenius, K. & Pedersen, A. G. Protein evolution is faster outside the cell. Mol. Biol. Evol. 23, 2039–2048 (2006).
Linksvayer, T. A. & Wade, M. J. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696 (2009).
Bensasson, D., Zarowiecki, M., Burt, A. & Koufopanou, V. Rapid evolution of yeast centromeres in the absence of drive. Genetics 178, 2161–2167 (2008).
Shi, J. et al. Widespread gene conversion in centromere cores. PLoS Biol. 8, e1000327 (2010).
Tirosh, I., Barkai, N. & Verstrepen, K. Promoter architecture and the evolvability of gene expression. J. Biol. 8, 95 (2009).
Carroll, S. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (2008).
West-Eberhard, M. J. Developmental Plasticity and Evolution (Oxford Univ. Press, USA, 2003).
Couzin, I. D., Krause, J., James, R., Ruxton, G. D. & Franks, N. R. Collective memory and spatial sorting in animal groups. J. Theor. Biol. 218, 1–11 (2002).
Sumpter, D. J. T. Collective Animal Behavior (Princeton Univ. Press, 2010).
Ben-Zvi, D. & Barkai, N. Scaling of morphogen gradients by an expansion-repression integral feedback control. Proc. Natl Acad. Sci. USA 107, 6924–6929 (2010).
Suel, G. M., Garcia-Ojalvo, J., Liberman, L. M. & Elowitz, M. B. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440, 545–550 (2006).
Xavier, J. B., Martinez-Garcia, E. & Foster, K. R. Social evolution of spatial patterns in bacterial biofilms: when conflict drives disorder. Am. Nat. 174, 1–12 (2009).
DeDeo, S., Krakauer, D. C. & Flack, J. C. Inductive game theory and the dynamics of animal conflict. PLoS Comput. Biol. 6, e1000782 (2010).
Frank, S. A. The Foundations of Social Evolution (Princeton Univ. Press, Princeton, New Jersey, 1998).
Wenseleers, T., Gardner, A. & Foster, K. R. in Social Behaviour: Genes, Ecology and Evolution (eds Szekely, T., Komdeur, J. & Moore, A. J.) 132–158 (Cambridge Univ. Press, 2010).
Rousset, F. Genetic Structure and Selection in Subdivided Populations (Princeton Univ. Press, 2004). This theory book shows the links among population genetics theory, multilevel selection theory, game theory, neighbour-modulated fitness theory and inclusive fitness theory.
Doebeli, M. & Hauert, C. Models of cooperation based on the prisoner's dilemma and the snowdrift game. Ecol. Lett. 8, 748–766 (2005).
Krebs, J. R. & Davies, N. B. An Introduction to Behavioural Ecology (Blackwell, Oxford, 1993).
Krieger, M. J. & Ross, K. G. Identification of a major gene regulating complex social behavior. Science 295, 328–332 (2002).
Wilson, D. S. Social semantics: toward a genuine pluralism in the study of social behaviour. J. Evol. Biol. 21, 368–373 (2008).
Foster, K. R. Diminishing returns in social evolution: the not-so-tragic commons. J. Evol. Biol. 17, 1058–1072 (2004).
Gardner, A. & Foster, K. R. in Ecology of Social Evolution (eds Korb, J. & Heinze, J.) 1–35 (Springer, Berlin, Heidelberg, 2008).
Beekman, M., Kondeur, J. & Ratnieks, F. L. W. Reproductive conflicts in social animals: who has power. Trends Ecol. Evol. 18, 277–282 (2003).
Smith, J. The social evolution of bacterial pathogenesis. Proc. Biol. Sci. 268, 61–69 (2001).
Boomsma, J. J. Lifetime monogamy and the evolution of eusociality. Phil. Trans. R. Soc. B 364, 3191–3207 (2009).
Alexander, R. D. The evolution of social behavior. Annu. Rev. Ecol. Syst. 5, 325–383 (1974).
Linksvayer, T. A. & Wade, M. J. The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: maternal effects, sib-social effects, and heterochrony. Q. Rev. Biol. 80, 317–336 (2005).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nature Rev. Microbiol. 3, 722–732 (2005).
Nowak, M. A., Tarnita, C. E. & Wilson, E. O. The evolution of eusociality. Nature 466, 1057–1062 (2010).
Wilson, D. S. & Wilson, E. O. Rethinking the theoretical foundation of sociobiology. Q. Rev. Biol. 82, 327–348 (2007).
Wilson, E. O. & Holldobler, B. Eusociality: origin and consequences. Proc. Natl Acad. Sci. USA 102, 13367–13371 (2005).
Krakauer, D. C. & Flack, J. C. Better living through physics. Nature 467, 661–661 (2010).
Dixon, T. The Invention of Altruism: Making Moral Meanings in Victorian Britain (Oxford Univ. Press for the British Academy, Oxford, 2008).
Ishii, S., Nishi, Y. & Egami, F. The fine structure of a pyocin. J. Mol. Biol. 13, 428–431 (1965)
Acknowledgements
Many thanks to W. Kim, S. Knowles, S. Mitri, G. Robinson, J. Schluter, O. Soyer, B. Stern, C. Thompson and K. Verstrepen for comments. I would also like to thank two referees for their comments on an earlier version of the paper, which were important in shaping the final presentation. K.R.F. is supported by European Research Council Grant 242670.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- Modern synthesis
-
The evolutionary synthesis in the early twentieth century that brought together Mendelian genetics with natural selection theory, giving birth to population genetics theory.
- Sociobiology
-
The study of social phenotypes, including both competitive and cooperative phenotypes.
- Sociogenomics
-
The study of the genomics and genetics of social behaviour. Key premises are that social behaviours have some genetic basis and that genes are functionally conserved across species.
- Inclusive fitness theory
-
Theory in which natural selection is analysed in terms of the effects of an actor on its own reproduction as well as its effects on other individuals; the latter effects are weighted by the genetic similarity (relatedness) between the actor and others.
- Multilevel selection theory
-
Theory in which natural selection is analysed in terms of the effects of an actor on its own reproduction as well as on members of its social group; the latter effects are weighted by the genetic similarity (relatedness) between the actor and the group.
- Evolutionary game theory
-
A theory designed to recognize frequency-dependent fitness effects in evolution, which occur because the strategies of one individual affect the fitness of another individual. Game theory logic is used in both inclusive fitness and multilevel selection.
- Functional scale
-
The number of genes, cells or organisms that operate towards a common evolutionary goal (the scale of agency). For example, the genes of a transposable element typically operate at a much lower functional scale than the genetic networks underlying cellular growth and division.
- Agent
-
A gene, cell, organism or any group of these that has the same evolutionary interests. Natural selection on agents leads to them to behave as though they are striving to maximize their genetic representation in later generations.
- Scale-free network
-
A network in which the distribution of the number of connections from each node follows a power law. This means that some nodes are highly connected (hubs), and paths through the network are shorter than those in highly ordered networks.
- Cheater mutant
-
A mutant that does not invest in a public good but benefits from the investment of others, such as a bacterial mutant that does not produce a secreted enzyme but uses the enzyme of wild-type cells.
- Eusocial insects
-
Social insects that display a division between work and reproduction among individuals. The clearest examples have morphologically distinct workers, as seen in many ants, bees, wasps and termites.
- Neighbour-modulated fitness theory
-
Closely aligned to inclusive fitness theory, this framework analyses natural selection in terms of the effects on the actor of its social trait, combined with the effects of the social trait in other individuals on the actor, where the latter effects are weighted by the genetic similarity (relatedness) between the actor and others.
- Social phenotype
-
A phenotype in one individual that affects the fitness of other individuals. 'Individual' here could mean a gene, cell, organism or even a group — whatever is appropriate for the analysis of the phenotype.
Rights and permissions
About this article
Cite this article
Foster, K. The sociobiology of molecular systems. Nat Rev Genet 12, 193–203 (2011). https://doi.org/10.1038/nrg2903
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2903
This article is cited by
-
When the “satisficing” is the new “fittest”: how a proscriptive definition of adaptation can change our view of cognition and culture
The Science of Nature (2022)
-
Genetic conflicts and the case for licensed anthropomorphizing
Behavioral Ecology and Sociobiology (2022)
-
Enforcement is central to the evolution of cooperation
Nature Ecology & Evolution (2019)
-
The meaning of intragenomic conflict
Nature Ecology & Evolution (2017)
-
Extinction, coexistence, and localized patterns of a bacterial population with contact-dependent inhibition
BMC Systems Biology (2014)