Key Points
-
Meiotic recombination events are distributed nonrandomly across the genome.
-
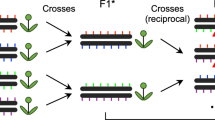
Single-molecule methods for the recovery of recombinant DNA molecules directly from sperm DNA have allowed recombination analysis to be carried out at high resolution in mammals.
-
Recent studies indicate that most meiotic recombination events (in both yeast and mammals) occur at highly localized hot spots (1–2 kb in width), whereas the bulk of the DNA is 'cold'.
-
Recombination hot spots in mice and humans are sites of initiation and resolution of both crossovers and non-crossover gene conversions.
-
Studies in model organisms, primarily budding yeast, provide direct insight into the molecular mechanisms behind hot-spot activity in mammals.
-
The primary determinant of the recombination distribution in yeast is the distribution of the double-strand breaks (DSBs) that initiate recombination. Recombination hot spots are DSB hot spots.
-
The frequency of DSBs in yeast is determined by the large-scale structural features of a chromosome as well as by local chromatin structure.
-
In principle, regional variation in crossover frequencies might also reflect differences in the likelihood that a given DSB will give rise to a crossover as opposed to a non-crossover product.
-
The fact that recombination hot spots are highly localized and separated by recombinationally 'cold' DNA is important in structuring the genome into linkage disequilibrium (LD)/haplotype blocks.
Abstract
Until recently, recombination studies in humans and mice had identified only a few anecdotal examples of crossover hot spots. Recently, the pace of discovery has accelerated. In every genomic segment that has been examined at sufficiently high resolution, recombination events have a punctate recombination distribution: they are clustered within small (1–2-kb) regions that are surrounded by large stretches of recombinationally suppressed DNA. Here, we review progress in understanding the distribution of mammalian recombination events, tie mammalian results together with informative studies in budding yeast and discuss the consequences of these findings for genome diversity and evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Petronczki, M., Siomos, M. F. & Nasmyth, K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423–440 (2003).
Moore, D. P. & Orr-Weaver, T. L. Chromosome segregation during meiosis: building an unambivalent bivalent. Curr. Top. Dev. Biol. 37, 263–299 (1998).
Lichten, M. & Goldman, A. S. Meiotic recombination hotspots. Annu. Rev. Genet. 29, 423–444 (1995). A lucid and detailed review of fungal and other recombination hot spots in meiosis.
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Hubert, R., MacDonald, M., Gusella, J. & Arnheim, N. High resolution localization of recombination hot spots using sperm typing. Nature Genet. 7, 420–424 (1994).
Arnheim, N., Calabrese, P. & Nordborg, M. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73, 5–16 (2003).
Jeffreys, A. J., Murray, J. & Neumann, R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell 2, 267–273 (1998).
Schneider, J. A., Peto, T. E., Boone, R. A., Boyce, A. J. & Clegg, J. B. Direct measurement of the male recombination fraction in the human β-globin hot spot. Hum. Mol. Genet. 11, 207–215 (2002).
Cullen, M., Perfetto, S. P., Klitz, W., Nelson, G. & Carrington, M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71, 759–776 (2002).
Jeffreys, A. J., Ritchie, A. & Neumann, R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet. 9, 725–733 (2000).
Jeffreys, A. J., Kauppi, L. & Neumann, R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 29, 217–222 (2001). Demonstrates the coincidence of linkage-disequilibrium breaks and sperm-crossover hot spots in the human major histocompatibility complex, and shows that most recombination events are highly localized.
May, C. A., Shone, A. C., Kalaydjieva, L., Sajantila, A. & Jeffreys, A. J. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nature Genet. 31, 272–275 (2002).
Lien, S., Szyda, J., Schechinger, B., Rappold, G. & Arnheim, N. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 66, 557–566 (2000).
Guillon, H. & de Massy, B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nature Genet. 32, 296–299 (2002).
Yauk, C. L., Bois, P. R. & Jeffreys, A. J. High-resolution sperm typing of meiotic recombination in the mouse MHC E β gene. EMBO J. 22, 1389–1397 (2003).
Thuriaux, P. Is recombination confined to structural genes on the eukaryotic genome? Nature 268, 460–462 (1977).
Baudat, F. & Nicolas, A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl Acad. Sci. USA 94, 5213–5218 (1997). With reference 28, this paper demonstrates the nonrandom distribution of double-strand breaks across large regions of the yeast genome.
Jeffreys, A. J. & May, C. A. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nature Genet. 36, 151–156 (2004). This paper and reference 14 demonstrate that mammalian sperm-crossover hot spots are also hot spots for non-crossover gene conversion.
Jeffreys, A. J. & May, C. A. DNA enrichment by allele-specific hybridization (DEASH): a novel method for haplotyping and for detecting low-frequency base substitutional variants and recombinant DNA molecules. Genome Res. 13, 2316–2324 (2003).
Qin, J., Richardson, L. L., Jasin, M., Handel, M. A. & Arnheim, N. Mouse strains with an active H2-Ea meiotic recombination hot spot exhibit increased levels of H2-Ea-specific DNA breaks in testicular germ cells. Mol. Cell. Biol. 24, 1655–1666 (2004).
Keeney, S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53 (2001).
Paques, F. & Haber, J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 (1999).
Smith, K. N. & Nicolas, A. Recombination at work for meiosis. Curr. Opin. Genet. Dev. 8, 200–211 (1998).
Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Romanienko, P. J. & Camerini-Otero, R. D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000).
Wu, T. -C. & Lichten, M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263, 515–518 (1994). This work provides one of the first and clearest descriptions of the link between double-strand break formation and local chromatin structure in yeast.
Zenvirth, D. et al. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 11, 3441–3447 (1992).
Gerton, J. L. et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 97, 11383–11390 (2000).
Fan, Q. Q. & Petes, T. D. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2037–2043 (1996).
Ohta, K., Shibata, T. & Nicolas, A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 13, 5754–5763 (1994).
Xu, L. & Kleckner, N. Sequence nonspecific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 14, 5115–5128 (1995).
Xu, F. & Petes, T. D. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics 143, 1115–1125 (1996).
Liu, J., Wu, T. -C. & Lichten, M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14, 4599–4608 (1995).
de Massy, B., Rocco, V. & Nicolas, A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14, 4589–4598 (1995).
Diaz, R. L., Alcid, A. D., Berger, J. M. & Keeney, S. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol. Cell. Biol. 22, 1106–1115 (2002).
Nicolas, A. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl Acad. Sci. USA 95, 87–89 (1998).
Wahls, W. P. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37, 37–75 (1998).
Petes, T. D. Meiotic recombination hot spots and cold spots. Nature Rev. Genet. 2, 360–369 (2001).
de Massy, B. Distribution of meiotic recombination sites. Trends Genet. 19, 514–522 (2003).
Kong, A. et al. A high-resolution recombination map of the human genome. Nature Genet. 31, 241–247 (2002).
Wu, T. -C. & Lichten, M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140, 55–66 (1995).
Borde, V., Wu, T. -C. & Lichten, M. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4832–4842 (1999). Demonstrates that recombination activity is partially a function of position on the chromosome.
Blat, Y., Protacio, R. U., Hunter, N. & Kleckner, N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802 (2002).
Ashley, T. G-band position effects on meiotic synapsis and crossing over. Genetics 118, 307–317 (1988).
Holmquist, G. P. Chromosome bands, their chromatin flavors, and their functional features. Am. J. Hum. Genet. 51, 17–37 (1992).
Kirkpatrick, D. T., Wang, Y. H., Dominska, M., Griffith, J. D. & Petes, T. D. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol. Cell. Biol. 19, 7661–7671 (1999).
Keeney, S. & Kleckner, N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells 1, 475–489 (1996).
White, M. A., Detloff, P., Strand, M. & Petes, T. D. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr. Genet. 21, 109–116 (1992).
Cao, L., Alani, E. & Kleckner, N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089–1101 (1990).
Nasar, F., Jankowski, C. & Nag, D. K. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol. 20, 3449–3458 (2000).
Jankowski, C., Nasar, F. & Nag, D. K. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA 97, 2134–2139 (2000).
Nag, D. K. & Kurst, A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics 146, 835–847 (1997).
White, M. A., Dominska, M. & Petes, T. D. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 90, 6621–6625 (1993).
Fan, Q., Xu, F. & Petes, T. D. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15, 1679–1688 (1995).
Kirkpatrick, D. T., Fan, Q. & Petes, T. D. Maximal stimulation of meiotic recombination by a yeast transcription factor requires the transcription activation domain and a DNA-binding domain. Genetics 152, 101–115 (1999).
Moens, P. B. et al. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106, 207–215 (1997).
Terasawa, M., Shinohara, A., Hotta, Y., Ogawa, H. & Ogawa, T. Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev. 9, 925–934 (1995).
Moens, P. B. et al. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622 (2002).
Fogel, S., Mortimer, R. K. & Lusnak, K. in The Molecular Biology of the Yeast Saccharomyces (eds. Strathern, J. N., Jones, E. W. & Broach, J. R.) 289–339 (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1981).
Mohrenweiser, H. W., Tsujimoto, S., Gordon, L. & Olsen, A. S. Regions of sex-specific hypo- and hyper-recombination identified through integration of 180 genetic markers into the metric physical map of human chromosome 19. Genomics 47, 153–162 (1998).
Yu, A. et al. Comparison of human genetic and sequence-based physical maps. Nature 409, 951–953 (2001).
Roberts, P. A. Differences in synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics 71, 401–415 (1972).
Parker, J. S., Palmer, R. W., Whitehorn, M. A. F. & Edgar, L. A. Chiasma frequency effects of structural chromosome change. Chromosoma 85, 673–686 (1982).
Hunter, N., Chambers, S. R., Louis, E. J. & Borts, R. H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15, 1726–1733 (1996).
Rayssiguier, C., Thaler, D. S. & Radman, M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342, 396–401 (1989).
Wall, J. D., Frisse, L. A., Hudson, R. R. & Di Rienzo, A. Comparative linkage-disequilibrium analysis of the β-globin hotspot in primates. Am. J. Hum. Genet. 73, 1330–1340 (2003).
Lamb, N. E. et al. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum. Mol. Genet. 6, 1391–1399 (1997).
Lamb, N. E. et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nature Genet. 14, 400–405 (1996).
Daly, M. J., Rioux, J. D., Schaffner, S. F., Hudson, T. J. & Lander, E. S. High-resolution haplotype structure in the human genome. Nature Genet. 29, 229–232 (2001).
Patil, N. et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294, 1719–1723 (2001).
Gabriel, S. B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002). With reference 70, this paper demonstrates that a large portion of the human genome is contained within haplotype blocks and that these blocks show limited diversity.
Phillips, M. S. et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nature Genet. 33, 382–387 (2003).
Wall, J. D. & Pritchard, J. K. Haplotype blocks and linkage disequilibrium in the human genome. Nature Rev. Genet. 4, 587–597 (2003).
Wang, N., Akey, J. M., Zhang, K., Chakraborty, R. & Jin, L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 71, 1227–1234 (2002).
Zhang, K. et al. Randomly distributed crossovers may generate block-like patterns of linkage disequilibrium: an act of genetic drift. Hum. Genet. 113, 51–59 (2003).
Anderson, E. C. & Slatkin, M. Population-genetic basis of haplotype blocks in the 5q31 region. Am. J. Hum. Genet. 74, 40–49 (2004).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
Stumpf, M. P. & McVean, G. A. Estimating recombination rates from population-genetic data. Nature Rev. Genet. 4, 959–968 (2003).
Kauppi, L., Sajantila, A. & Jeffreys, A. J. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum. Mol. Genet. 12, 33–40 (2003).
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F. W. The double-strand-break repair model for recombination. Cell 33, 25–35 (1983).
Nicolas, A., Treco, D., Schultes, N. P. & Szostak, J. W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338, 35–39 (1989). This is a reader-friendly description of a meiotic conversion hot spot in yeast that is accessible to the non-specialist.
Jeffreys, A. J. & Neumann, R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nature Genet. 31, 267–271 (2002).
Boulton, A., Myers, R. S. & Redfield, R. J. The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl Acad. Sci. USA 94, 8058–8063 (1997).
Ardlie, K. G., Kruglyak, L. & Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nature Rev. Genet. 3, 299–309 (2002).
Nachman, M. W. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17, 481–485 (2001).
Strathern, J. N., Shafer, B. K. & McGill, C. B. DNA synthesis errors associated with double-strand-break repair. Genetics 140, 965–972 (1995).
Cullen, M. et al. Characterization of recombination in the HLA class II region. Am. J. Hum. Genet. 60, 397–407 (1997).
Tease, C., Hartshorne, G. M. & Hulten, M. A. Patterns of meiotic recombination in human fetal oocytes. Am. J. Hum. Genet. 70, 1469–1479 (2002).
Lynn, A. et al. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296, 2222–2225 (2002).
Mitra, R. D. et al. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl Acad. Sci. USA 100, 5926–5931 (2003).
Hattori, M. et al. The DNA sequence of human chromosome 21. Nature 405, 311–319 (2000).
Broman, K. W., Murray, J. C., Sheffield, V. C., White, R. L. & Weber, J. L. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63, 861–869 (1998).
Lynn, A. et al. Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res. 10, 1319–1332 (2000).
Anderson, L. K., Reeves, A., Webb, L. M. & Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579 (1999).
Barlow, A. L. & Hulten, M. A. Crossing over analysis at pachytene in man. Eur. J. Hum. Genet. 6, 350–358 (1998).
Froenicke, L., Anderson, L. K., Wienberg, J. & Ashley, T. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71, 1353–1368 (2002).
Sun, F. et al. Human male recombination maps for individual chromosomes. Am. J. Hum. Genet. 74, 521–531 (2004).
Laurie, D. A. & Hulten, M. A. Further studies on chiasma distribution and interference in the human male. Ann. Hum. Genet. 49, 203–214 (1985).
Laurie, D. A. & Hulten, M. A. Further studies on bivalent chiasma frequency in human males with normal karyotypes. Ann. Hum. Genet. 49, 189–201 (1985).
Lawrie, N. M., Tease, C. & Hulten, M. A. Chiasma frequency, distribution and interference maps of mouse autosomes. Chromosoma 104, 308–314 (1995).
Shiroishi, T., Koide, T., Yoshino, M., Sagai, T. & Moriwaki, K. Hotspots of homologous recombination in mouse meiosis. Adv. Biophys. 31, 119–132 (1995).
Hedrick, P. W. Inference of recombinational hotspots using gametic disequilibrium values. Heredity 60, 435–438 (1988).
Chakravarti, A. et al. Nonuniform recombination within the human β-globin gene cluster. Am. J. Hum. Genet. 36, 1239–1258 (1984).
Smith, R. A., Ho, P. J., Clegg, J. B., Kidd, J. R. & Thein, S. L. Recombination breakpoints in the human β-globin gene cluster. Blood 92, 4415–4421 (1998).
Yip, S. P., Lovegrove, J. U., Rana, N. A., Hopkinson, D. A. & Whitehouse, D. B. Mapping recombination hotspots in human phosphoglucomutase (PGM1). Hum. Mol. Genet. 8, 1699–1706 (1999).
Cruciani, F. et al. Linkage disequilibrium analysis of the human adenosine deaminase (ada) gene provides evidence for a lack of correlation between hot spots of equal and unequal homologous recombination. Genomics 82, 20–33 (2003).
Twells, R. C. et al. Haplotype structure, LD blocks, and uneven recombination within the LRP5 gene. Genome Res. 13, 845–855 (2003).
Cullen, M., Erlich, H., Klitz, W. & Carrington, M. Molecular mapping of a recombination hotspot located in the second intron of the human TAP2 locus. Am. J. Hum. Genet. 56, 1350–1358 (1995).
Bryda, E. C., DePari, J. A., Sant'Angelo, D. B., Murphy, D. B. & Passmore, H. C. Multiple sites of crossing over within the Eb recombinational hotspot in the mouse. Mamm. Genome 2, 123–129 (1992).
Lafuse, W. P. & David, C. S. Recombination hot spots within the I region of the mouse H-2 complex map to the E β and E α genes. Immunogenetics 24, 352–360 (1986).
Zimmerer, E. J. & Passmore, H. C. Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics 33, 132–140 (1991).
Uematsu, Y., Kiefer, H., Schulze, R., Fischer-Lindahl, K. & Steinmetz, M. Molecular characterization of a meiotic recombinational hotspot enhancing homologous equal crossing-over. EMBO J. 5, 2123–2129 (1986).
Steinmetz, M., Stephan, D. & Fischer Lindahl, K. Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell 44, 895–904 (1986). Provides the first account of localized crossover clustering in mammalian pedigrees.
Shiroishi, T. et al. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics 31, 79–88 (1990).
Snoek, M., Teuscher, C. & van Vugt, H. Molecular analysis of the major MHC recombinational hot spot located within the G7c gene of the murine class III region that is involved in disease susceptibility. J. Immunol. 160, 266–272 (1998).
Mizuno, K., Koide, T., Sagai, T., Moriwaki, K. & Shiroishi, T. Molecular analysis of a recombinational hotspot adjacent to Lmp2 gene in the mouse MHC: fine location and chromatin structure. Mamm. Genome 7, 490–496 (1996).
Shiroishi, T., Sagai, T., Hanzawa, N., Gotoh, H. & Moriwaki, K. Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J. 10, 681–686 (1991). Demonstrates the effect of modifying factors on crossing-over at a localized hot spot.
Laan, M. & Paabo, S. Demographic history and linkage disequilibrium in human populations. Nature Genet. 17, 435–438 (1997).
Zavattari, P. et al. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum. Mol. Genet. 9, 2947–2957 (2000).
Hedrick, P. W. Gametic disequilibrium measures: proceed with caution. Genetics 117, 331–341 (1987).
Lewontin, R. C. The interaction of selection and linkage. II. Optimum models. Genetics 50, 757–782 (1964).
Weiss, K. M. & Clark, A. G. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18, 19–24 (2002).
Gray, I. C., Campbell, D. A. & Spurr, N. K. Single nucleotide polymorphisms as tools in human genetics. Hum. Mol. Genet. 9, 2403–2408 (2000).
Johnson, G. C. et al. Haplotype tagging for the identification of common disease genes. Nature Genet. 29, 233–237 (2001).
National Human Genome Research Institute. International consortium launches genetic variation mapping project. NIH News Advisory [online], <http://genome.gov/10005336> (2002).
Yao, H. et al. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl Acad. Sci. USA 99, 6157–6162 (2002).
Game, J. C., Sitney, K. C., Cook, V. E. & Mortimer, R. K. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand breaks, and sister-chromatid exchange in yeast. Genetics 123, 695–713 (1989).
Sun, H., Treco, D., Schultes, N. P. & Szostak, J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338, 87–90 (1989).
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). With reference 130, this paper identifies Spo11 as the catalytic subunit of the meiotic double-strand break machinery, and provides evidence that it cuts DNA through a topoisomerase-like transesterase reaction.
Bergerat, A. et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997).
Sun, H., Treco, D. & Szostak, J. W. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64, 1155–1161 (1991).
Bishop, D. K., Park, D., Xu, L. & Kleckner, N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456 (1992).
Shinohara, A., Ogawa, H. & Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470 (1992).
Yuhki, N. et al. Comparative genome organization of human, murine, and feline MHC class II region. Genome Res. 13, 1169–1179 (2003).
Isobe, T. et al. Molecular characterization of the Pb recombination hotspot in the mouse major histocompatibility complex class II region. Genomics 80, 229–235 (2002).
Acknowledgements
L.K. was supported in part by the Instrumentarium Science Foundation, the Finnish Cultural Foundation and the Osk. Huttunen Foundation. A.J.J. is supported by grants from the Royal Society, the Medical Research Council and the Wellcome Trust (UK). S.K. is supported by grants from the US National Institutes of Health and the Byrne Fund.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez
FURTHER INFORMATION
GermOnline (a web database for information about germ cell biology, including meiosis)
Glossary
- HAPLOTYPE
-
A combination of alleles at different loci that is transmitted together from one generation to the next.
- CENTIMORGAN
-
(cM). A measure of genetic distance. 1 cM corresponds to a 1% frequency of recombinant progeny.
- CHIASMA
-
(pl. chiasmata). A cytologically visible physical connection between homologous chromosomes that corresponds to the position of a meiotic crossover.
- LINKAGE DISEQUILIBRIUM
-
(LD). A measure of whether alleles at two loci coexist in a population in a nonrandom fashion. Alleles that are in LD are found together on the same haplotype more often than would be expected by chance.
- MINISATELLITES
-
Regions of DNA in which repeat units of 6–100 bp are arranged in tandem arrays that are 0.5–30 kb in length.
- LD BLOCK
-
A DNA segment within which markers are in significant linkage disequilibrium with each other, which implies that there is low recombination activity within the block.
- GENE CONVERSION
-
The non-reciprocal transfer of information between homologous DNA sequences as a consequence of heteroduplex formation during recombination, which is followed by repair of mismatches in the heteroduplex.
- ADMIXTURE
-
The mixing of two genetically differentiated populations.
- ENRICHMENT-BASED ASSAY
-
An allele-specific hybridization method for enriching DNA molecules that carry certain allelic combinations from bulk genomic DNA.
- LIKELIHOOD RATIO
-
The relative likelihood of obtaining observed experimental data under two different models.
- ISOCHORE
-
A region of genomic DNA sequence in which G+C compositions are relatively uniform.
- R-BANDS
-
A chromosome banding pattern that is produced with various staining procedures; it is reciprocal to the pattern that is produced by Giemsa staining (G-Bands).
- HOLLIDAY JUNCTION
-
A point at which the strands of two double-stranded DNA molecules exchange partners, which occurs as an intermediate in crossing-over.
- COALESCENT APPROACHES
-
A means of investigating the shared genealogical history of genes. A genealogy is constructed backwards in time starting with the present-day sample. Lineages coalesce when they have a common ancestor.
- MEIOTIC DRIVE
-
Distortion of meiotic inheritance such that one allele at a heterozygous site is recovered in greater than half of the gametes (as opposed to the expected 50:50 Mendelian segregation).
- PCR COLONY METHOD
-
A method in which individual DNA molecules are immobilized in a matrix and then subjected to PCR amplification in situ. For (haploid) sperm DNA, polymorphic sites can be typed within amplification products to yield haplotypes of chromosomal regions, allowing, in principle, the identification of recombinant molecules.
Rights and permissions
About this article
Cite this article
Kauppi, L., Jeffreys, A. & Keeney, S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet 5, 413–424 (2004). https://doi.org/10.1038/nrg1346
Issue Date:
DOI: https://doi.org/10.1038/nrg1346
This article is cited by
-
A loss-of-function variant in ZCWPW1 causes human male infertility with sperm head defect and high DNA fragmentation
Reproductive Health (2024)
-
Molecular mechanisms and regulation of recombination frequency and distribution in plants
Theoretical and Applied Genetics (2024)
-
Removal of a partial genomic duplication restores synaptic transmission and behavior in the MyosinVA mutant mouse Flailer
BMC Biology (2023)
-
Genome-wide recombination map construction from single sperm sequencing in cattle
BMC Genomics (2022)
-
The histone modification reader ZCWPW1 promotes double-strand break repair by regulating cross-talk of histone modifications and chromatin accessibility at meiotic hotspots
Genome Biology (2022)