Key Points
-
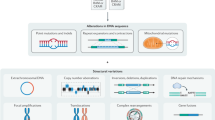
The genomics revolution has catalysed the development of new technologies that can be applied to provide a comprehensive view of the molecular changes that occur during cancer development.
-
Three independent projects — the Cancer Genome Anatomy Project (CGAP), the Human Cancer Genome Project (HCGP) and the Cancer Genome Project (CGP) — have applied sequence-based technologies to produce synergistic data sets that are amenable to integration.
-
The data of these projects are derived from the human genome (through sequencing of gene exons to identify cancer mutations), as well as from the human transcriptome in the form of expressed sequence tags (ESTs) and serial analysis of gene expression (SAGE) tags that are generated from tumours and normal tissues.
-
The CGAP has facilitated the interface of the human genome sequence with the cytogenetic map through FISH-mapping of BAC clones that were substrates for generating the finished genome sequence. This linkage facilitates the characterization of chromosomal aberrations that are associated with cancer.
-
A suite of informatics tools is accessible through the CGAP website that allow in silico analysis of CGAP and HCGP gene-expression data, polymorphisms and chromosomal aberrations of cancer. In the future, these data sets will be integrated with the mutation analysis of the CGP.
-
The data sets that are generated by these projects are a platform for a variety of applications in cancer research, such as the design and generation of microarrays.
Abstract
Technologies that provide a genome-wide view offer an unprecedented opportunity to scrutinize the molecular biology of the cancer cell. The information that is derived from these technologies is well suited to the development of public databases of alterations in the cancer genome and its expression. Here, we describe the synergistic efforts of research programmes in Brazil, the United Kingdom and the United States towards building integrated databases that are widely accessible to the research community, to enable basic and applied applications in cancer research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Van Dyke, T. & Jacks, T. Cancer modeling in the modern era: progress and challenges. Cell 108, 135–144 (2002).
Dunn, G. P., Bruce, A. T., Ikeda, H., Old, L. J. & Schreiber, R. D. Cancer immunoediting: from immunosurveillance to tumor escape. Nature Immunol. 3, 991–998 (2002).
Adams, M. D. et al. Sequence identification of 2,375 human brain genes. Nature 355, 632–634 (1992).The development and early application of ESTs to study human gene expression.
Strausberg, R. L. & Riggins, G. J. Navigating the human transcriptome. Proc. Natl Acad. Sci. USA 98, 11837–11838 (2001).
Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. Serial analysis of gene-expression. Science 270, 484–487 (1995).The initial description of the serial analysis of gene-expression strategy.
Strausberg, R. L., Buetow, K. H., Emmert-Buck, M. R. & Klausner, R. D. The cancer genome anatomy project — building an annotated gene index. Trends Genet. 16, 103–106 (2000).The launch of the Cancer Genome Anatomy Project.
Riggins, G. J. et al. SAGEmap: a gene expression resource for the Cancer Genome Anatomy Project. Am. J. Human Genet. 67, 357 (2000).
Boon, K. et al. An anatomy of normal and malignant gene expression. Proc. Natl Acad. Sci. USA 99, 11287–11292 (2002).
Hough, C. D. et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 60, 6281–6287 (2000).
Loging, W. T. et al. Identifying potential tumor markers and antigens by database mining and rapid expression screening. Genome Res. 10, 1393–1402 (2000).
Riggins, G. J. Using serial analysis of gene expression to identify tumor markers and antigens. Dis. Markers 17, 41–48 (2001).
Porter, D. A. et al. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 61, 5697–5702 (2001).
St Croix, B. et al. Genes expressed in human tumor endothelium. Science 289, 1197–1202 (2000).
Lal, A. et al. Transcriptional response to hypoxia in human tumors. J. Natl Cancer Inst. 93, 1337–1343 (2001).
Birney, E., Clamp, M. & Hubbard, T. Databases and tools for browsing genomes. Annu. Rev. Genomics Hum. Genet. 3, 293–310 (2002).
Hubbard, T. et al. The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 (2002).Ensembl combines many genomic data sources to provide a comprehensive view of the human and other genomes. In the future, cancer genome data could be viewed in a similar way.
Karolchik, D. et al. The UCSC genome browser database. Nucleic Acids Res. 31, 51–54 (2003).
Druker, B. J. Perspectives on the development of a molecularly targeted agent. Cancer Cell 1, 31–36 (2002).
Cheung, V. G. et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409, 953–958 (2001).
Kirsch, I. R. & Ried, T. Integration of cytogenetic data with genome maps and available probes: present status and future promise. Semin. Hematol. 37, 420–428 (2000).
Kirsch, I. R. et al. A systematic, high-resolution linkage of the cytogenetic and physical maps of the human genome. Nature Genet. 24, 339–340 (2000).
Schaefer, C., Grouse, L., Buetow, K. & Strausberg, R. L. A new cancer genome anatomy project web resource for the community. Cancer J. 7, 52–60 (2001).
Neto, E. D. et al. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA 97, 3491–3496 (2000).The development and application of ORESTES.
Neto, E. D. et al. Mini-libraries constructed from cDNA generated by arbitrarily primed RT-PCR: an alternative to normalized libraries for the generation of ESTs from nanogram quantities of mRNA. Gene 186, 135–142 (1997).
de Souza, S. J. et al. Identification of human chromosome 22 transcribed sequences with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA 97, 12690–12693 (2000).
Camargo, A. A. The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc. Natl Acad. Sci. USA 98, 12103–12108 (2001).
The International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Quackenbush, J. et al. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29, 159–164 (2001).
Christoffels, A. et al. STACK: sequence tag alignment and consensus knowledgebase. Nucleic Acids Res. 29, 234–238 (2001).
Wheeler, D. L. et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 31, 28–33 (2003).
Reymond, A. et al. Nineteen additional unpredicted transcripts from human chromosome 21. Genomics 79, 824–832 (2002).
Rondeau, G. et al. Comprehensive analysis of a large genomic sequence at the putative B-cell chronic lymphocytic leukaemia (B-CLL) tumour suppresser gene locus. Mutat. Res. 458, 55–70 (2001).
Bullrich, F. et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 61, 6640–6648 (2001).
Montpetit, A., Boily, G. & Sinnett, D. A detailed transcriptional map of the chromosome 12p12 tumour suppressor locus. Eur. J. Hum. Genet. 10, 62–71 (2002).
Sood, R. et al. Cloning and characterization of 13 novel transcripts and the human RC58 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics 73, 211–222 (2001).
Buetow, K. H. et al. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA 98, 581–584 (2001).
Mironov, A. A., Fickett, J. W. & Gelfand, M. S. Frequent alternative splicing of human genes. Genome Res. 9, 1288–1293 (1999).
Modrek, B. & Lee, C. A genomic view of alternative splicing. Nature Genet. 30, 13–19 (2002).
Xu, Q., Modrek, B. & Lee, C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 30, 3754–3766 (2002).
Xie, H. et al. Computational analysis of alternative splicing using EST tissue information. Genomics 80, 326 (2002).
Correa, R. G., de Carvalho, A. F., Pinheiro, N. A., Simpson, A. J. G. & de Souza, S. J. NABC1 (BCAS1): alternative splicing and downregulation in colorectal tumors. Genomics 65, 299–302 (2000).
Iseli, C. et al. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 12, 1068–1074 (2002).
Strausberg, R. L., Buetow, K. H., Greenhut, S. F., Grouse, L. H. & Schaefer, C. F. The cancer genome anatomy project: online resources to reveal the molecular signatures of cancer. Cancer Invest. 20, 1038–1050 (2002).
Strausberg, R. L., Greenhut, S. F., Grouse, L. H., Schaefer, C. F. & Buetow, K. H. In silico analysis of cancer through the cancer genome anatomy project. Trends Cell Biol. 11, 66–71 (2001).
Leerkes, M. R. et al. In silico comparison of the transcriptome derived from purified normal breast cells and breast tumor cell lines reveals candidate upregulated genes in breast tumor cells. Genomics 79, 257–265 (2002).
Schmitt, A. O. et al. Exhaustive mining of EST libraries for genes differentially expressed in normal and tumour tissues. Nucleic Acids Res. 27, 4251–4260 (1999).
Mitas, M. et al. Prostate-specific Ets (PSE) factor: a novel marker for detection of metastatic breast cancer in axillary lymph nodes. Br. J. Cancer 86, 899–904 (2002).
Olsson, P. et al. GDEP, a new gene differentially expressed in normal prostate and prostate cancer. Prostate 48, 231–241 (2001).
Nelson, P. S. et al. Comprehensive analyses of prostate gene expression: convergence of expressed sequence tag databases, transcript profiling and proteomics. Electrophoresis 21, 1823–1831 (2000).
Nelson, P. S. Identifying immunotherapeutic targets for prostate carcinoma through the analysis of gene expression profiles. Ann. NY Acad. Sci. 975, 232–245 (2002).
De Young, M. P., Damania, H., Scheurle, D., Zylberberg, C. & Narayanan, R. Bioinformatics-based discovery of a novel factor with apparent specificity to colon cancer. In Vivo 16, 239–248 (2002).
Shillitoe, E. J. et al. Genome-wide analysis of oral cancer — early results from the Cancer Genome Anatomy Project. Oral Oncol. 36, 8–16 (2000).
Patel, V., Leethanakul, C. & Gutkind, J. S. New approaches to the understanding of the molecular basis of oral cancer. Crit. Rev. Oral Biol. Med. 12, 55–63 (2001).
Brinkmann, U. et al. PAGE-1, an X chromosome-linked GAGE-like gene that is expressed in normal and neoplastic prostate, testis, and uterus. Proc. Natl Acad. Sci. USA 95, 10757–10762 (1998).
Vinals, C., Gaulis, S. & Coche, T. Using in silico transcriptomics to search for tumor-associated antigens for immunotherapy. Vaccine 19, 2607–2614 (2001).
Scanlan, M. J. et al. Identification of cancer/testis genes by database mining and mRNA expression analysis. Int. J. Cancer 98, 485–492 (2002).An excellent example of the use of the EST databases to identify genes that are relevant to cancer.
Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).The first report of mutations in the BRAF gene.
Lowinger, T. B., Riedl, B., Dumas, J. & Smith, R. A. Design and discovery of small molecules targeting Raf-1 kinase. Curr. Pharm. Des. 8, 2269–2278 (2002).
Miller, D. G. On the nature of susceptibility to cancer. The presidential address. Cancer 46, 1307–1318 (1980).
Strausberg, R. L. et al. An international database and integrated analysis tools for the study of cancer gene expression. Pharmacogenomics J. 2, 156–164 (2002).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Chung, C. H., Bernard, P. S. & Perou, C. M. Molecular portraits and the family tree of cancer. Nature Genet. 32, 533–540 (2002).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. New Eng. J. Med. 347, 1999–2009 (2002).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Alizadeh, A. et al. The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harb. Symp. Quant. Biol. 64, 71–78 (1999).
Kapranov, P. et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296, 916–919 (2002).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Buetow, K. H. et al. Cancer Molecular Analysis Project: weaving a rich cancer research tapestry. Cancer Cell 1, 315–318 (2002).
Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. Serial analysis of gene-expression. Science 270, 484–487 (1995).
Druker, B. Signal transduction inhibition: results from phase I clinical trials in chronic myeloid leukemia. Semin. Hematol. 38, 9–14 (2001).A classic example of targeted molecular therapeutics.
Druker, B. J. Imatinib and chronic myeloid leukemia: validating the promise of molecularly targeted therapy. Eur. J. Cancer 38, 70–76 (2002).
Rozycka, M., Collins, N., Stratton, M. R. & Wooster, R. Rapid detection of DNA sequence variants by conformation-sensitive capillary electrophoresis. Genomics 70, 34–40 (2000).
Acknowledgements
R.L.S. thanks R. Klausner for initiating the Cancer Genome Anatomy Project and for vigorous encouragement and support during the implementation. A.J.G.S. is indebted to F. Perez and L. Old, respectively the Scientific Directors of the State of São Paulo Research Foundation (FAPESP) and the Ludwig Institute for Cancer Research, for their enthusiastic support of the Human Cancer Genome Project. R.W. thanks M. Stratton, A. Futreal and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
LocusLink
OMIM
FURTHER INFORMATION
Mitelman Database of Chromosome Aberrations in Cancer
Spectral Karyotyping/Comparative Genomic Hybridization Database
Glossary
- TRANSCRIPTOME
-
The complete catalogue of all the RNA species of a cell, tissue or organism.
- cDNA
-
Complementary DNA that is produced from an RNA template by an RNA-dependent DNA polymerase
- HYPOXIA
-
A physiological state in which insufficient oxygen reaches a tissue.
- FLUORESCENCE IN SITU HYBRIDIZATION
-
(FISH). A technology in which chromosomes (or chromosomal segments) are painted with fluorescent molecules.
- BACTERIAL ARTIFICIAL CHROMOSOME
-
(BAC). A DNA molecule that can be propagated in bacteria and is useful for cloning large (100–200 kb) segments of DNA from other species.
- PHASE I TRIAL
-
The first stage in a clinical trial, which is designed to assess the safety and dosage levels of a new treatment, and usually involves only a few patients.
- BLAST CRISIS
-
The progression of myeloid leukaemia from a clonal proliferation of myeloid cells to a highly refractory progressive disease with >30% blast cells in the peripheral blood and bone marrow, and a one-year survival of <10%.
- HETERODUPLEX ASSAY
-
A rapid method to detect mutations that relies on the fact that double-stranded DNA molecules with a single base-pair mismatch migrate to a different location compared with molecules that do not have a mismatch.
- PASSENGER ALTERATIONS
-
A phenomenon that refers to the fact that some mutations do not seem to have any functional benefit for the tumour; they probably arise owing to faulty DNA-repair machinery in the tumour, and are 'just along for the ride'.
- COMPARATIVE GENOMIC HYBRIDIZATION
-
(CGH). A technology through which tumour and reference DNA are differentially labelled to show copy-number changes in tumour genomes
- SPECTRAL KARYOTYPING
-
(SKY). A technique for painting each chromosome in a different colour, which is useful for looking at chromosomal aberrations such as translocations.
Rights and permissions
About this article
Cite this article
Strausberg, R., Simpson, A. & Wooster, R. Sequence-based cancer genomics: progress, lessons and opportunities. Nat Rev Genet 4, 409–418 (2003). https://doi.org/10.1038/nrg1085
Issue Date:
DOI: https://doi.org/10.1038/nrg1085
This article is cited by
-
Omics-based molecular techniques in oral pathology centred cancer: prospect and challenges in Africa
Cancer Cell International (2017)
-
Understanding cancer complexome using networks, spectral graph theory and multilayer framework
Scientific Reports (2017)
-
Predicting potential cancer genes by integrating network properties, sequence features and functional annotations
Science China Life Sciences (2013)
-
Suppression subtractive hybridization profiles of radial growth phase and metastatic melanoma cell lines reveal novel potential targets
BMC Cancer (2008)
-
In silico whole-genome screening for cancer-related single-nucleotide polymorphisms located in human mRNA untranslated regions
BMC Genomics (2007)