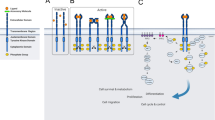
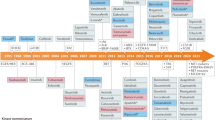
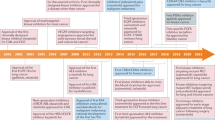
Abstract
Receptor tyrosine kinase signalling pathways have been successfully targeted to inhibit proliferation and angiogenesis for cancer therapy. However, kinase deregulation has been firmly demonstrated to play an essential role in virtually all major disease areas. Kinase inhibitor drug discovery programmes have recently broadened their focus to include an expanded range of kinase targets and therapeutic areas. In this Review, we provide an overview of the novel targets, biological processes and disease areas that kinase-targeting small molecules are being developed against, highlight the associated challenges and assess the strategies and technologies that are enabling efficient generation of highly optimized kinase inhibitors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
U.S. Food & Drug Administration. New drugs at FDA: CDER's new molecular entities and new therapeutic biological products. FDA https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/default.htm (2017).
Cohen, P. & Alessi, D. R. Kinase drug discovery — what's next in the field? ACS Chem. Biol. 8, 96–104 (2013).
Ficarro, S. B. et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 (2002).
Cohen, P. The regulation of protein function by multisite phosphorylation — a 25 year update. Trends Biochem. Sci. 25, 596–601 (2000).
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Muller, S., Chaikuad, A., Gray, N. S. & Knapp, S. The ins and outs of selective kinase inhibitor development. Nat. Chem. Biol. 11, 818–821 (2015).
Levitzki, A. Protein kinase inhibitors as a therapeutic modality. Acc. Chem. Res. 36, 462–469 (2003).
National Institutes of Health Office of Strategic Coordination - The Common Fund. Understudied proteins. NIH https://commonfund.nih.gov/idg/understudiedproteins (2017).
Fedorov, O., Muller, S. & Knapp, S. The (un)targeted cancer kinome. Nat. Chem. Biol. 6, 166–169 (2010).
Botta, M. New frontiers in kinases: special issue. ACS Med. Chem. Lett. 5, 270 (2014).
Zhao, Q. et al. Broad-spectrum kinase profiling in live cells with lysine-targeted sulfonyl fluoride probes. J. Am. Chem. Soc. 139, 680–685 (2017). This paper describes an in cell competition assay to discern kinase inhibitor selectivity in a physiological context.
Savitski, M. M. et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). This paper describes a proteome-wide profiling technique based on the CETSA assay.
Aldeghi, M., Heifetz, A., Bodkin, M. J., Knapp, S. & Biggin, P. C. Predictions of ligand selectivity from absolute binding free energy calculations. J. Am. Chem. Soc. 139, 946–957 (2017). This article describes an unusually accurate computational method to determine inhibitor selectivity.
Dale, T. et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat. Chem. Biol. 11, 973–980 (2015).
Alexander, P. B. & Wang, X. F. Resistance to receptor tyrosine kinase inhibition in cancer: molecular mechanisms and therapeutic strategies. Front. Med. 9, 134–138 (2015).
Villicana, C., Cruz, G. & Zurita, M. The basal transcription machinery as a target for cancer therapy. Cancer Cell. Int. 14, 18 (2014).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2017).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Kwiatkowski, N. et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014). This paper demonstrates effective targeting of transcriptional vulnerabilities in cancer via a covalent CDK7 kinase inhibitor.
Christensen, C. L. et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 (2014).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Bywater, M. J., Pearson, R. B., McArthur, G. A. & Hannan, R. D. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 13, 299–314 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Hann, S. R. & Eisenman, R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol. 4, 2486–2497 (1984).
Herrick, D. J. & Ross, J. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3' untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 14, 2119–2128 (1994).
Posternak, V. & Cole, M. D. Strategically targeting MYC in cancer. F1000Res. 5, 408 (2016).
Gonda, T. J. & Ramsay, R. G. Directly targeting transcriptional dysregulation in cancer. Nat. Rev. Cancer 15, 686–694 (2015).
Di Vona, C. et al. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol. Cell 57, 506–520 (2015).
Ionescu, A. et al. DYRK1A kinase inhibitors with emphasis on cancer. Mini Rev. Med. Chem. 12, 1315–1329 (2012).
Zhou, Y., Shen, J. K., Hornicek, F. J., Kan, Q. & Duan, Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget 7, 40846–40859 (2016).
Schachter, M. M. & Fisher, R. P. The CDK-activating kinase Cdk7: taking yes for an answer. Cell Cycle 12, 3239–3240 (2013).
Guo, J. & Price, D. H. RNA polymerase II transcription elongation control. Chem. Rev. 113, 8583–8603 (2013).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT03269669 (2018).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Pelish, H. E. et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526, 273–276 (2015). This paper describes a highly selective natural product inhibitor of CDK8, cortistatin A, which downregulates SE-associated genes in AML. This study also introduces the concept that cancer cells are highly sensitive to SE-controlled gene dosage; both upregulation and downregulation of these genes is toxic.
Clarke, P. et al. Discovery of preclinical development candidate inhibitors of the mediator complex-associated kinases CDK8 and CDK19 and evaluation of their therapeutic potential [abstract]. Cancer Res. 76 (Suppl.), 3025 (2016).
Bahr, B. L. et al. Combination strategies to target super enhancer transcriptional activity by CDK9 and BRD4 inhibition in acute myeloid leukemia [abstract]. Cancer Res. 75 (Suppl.), 2698 (2015).
Sonawane, Y. A. et al. Cyclin dependent kinase 9 inhibitors for cancer therapy. J. Med. Chem. 59, 8667–8684 (2016).
Yin, T. et al. B.; de Dios, A. ; Du, J., A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol. Cancer Ther. 13, 1442–1456 (2014).
Morales, F. & Giordano, A. Overview of CDK9 as a target in cancer research. Cell Cycle 15, 519–527 (2016).
Lu, H. et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. eLife 4, e06535 (2015).
Barsanti, P. A. et al. Pyridine and pyrazine derivatives as protein kinase modulators. US Patent WO2011012661 A1 (2010).
Liang, K. et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 35, 928–938 (2015).
Zhang, T. et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 12, 876–884 (2016). This paper describes an inhibitor that induces a loop rearrangement, enabling covalent inhibition of a distal cysteine by an ATP-competitive inhibitor.
Hamman, K. et al. Targeting the transcriptional kinases CDK12 and CDK13 in breast and ovarian cancer [abstract]. FASEB J. 31 (Suppl. 1), 938.9 (2017).
Johnson, S. F. et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 17, 2367–2381 (2016).
Bao, Z. et al. Effectiveness and safety of poly (ADP-ribose) polymerase inhibitors in cancer therapy: a systematic review and meta-analysis. Oncotarget 7, 7629–7639 (2016).
Vinay, D. S. et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 35 (Suppl.), S185–S198 (2015).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Sukari, A., Nagasaka, M., Al-Hadidi, A. & Lum, L. G. Cancer Immunology and immunotherapy. Anticancer Res. 36, 5593–5606 (2016).
Lemke, G. & Rothlin, C. V. Immunobiology of the TAM receptors. Nat. Rev. Immunol. 8, 327–336 (2008).
Seitz, H. M., Camenisch, T. D., Lemke, G., Earp, H. S. & Matsushima, G. K. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J. Immunol. 178, 5635–5642 (2007).
Caraux, A. et al. Natural killer cell differentiation driven by Tyro3 receptor tyrosine kinases. Nat. Immunol. 7, 747–754 (2006).
Zhang, Z. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 44, 852–860 (2012).
Postel-Vinay, S. & Ashworth, A. AXL and acquired resistance to EGFR inhibitors. Nat. Genet. 44, 835–836 (2012).
[No authors listed.] Clinical trials. BerGenBio http://www.bergenbio.com/pipeline/ongoing-clinical-trials/ (2017).
Myers, S. H., Brunton, V. G. & Unciti-Broceta, A. AXL inhibitors in cancer: a medicinal chemistry perspective. J. Med. Chem. 59, 3593–3608 (2016).
Paolino, M. et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 507, 508–512 (2014). This paper validates TAM kinases as therapeutic targets in cancer, demonstrating that their pharmacological inhibition enhances anti-metastatic NK cell activity.
Putz, E. M. et al. CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance. Cell Rep. 4, 437–444 (2013).
Johannessen, L. et al. Small-molecule studies identify CDK8 as a regulator of IL-10 in myeloid cells. Nature Chem. Biol. 13, 1102–1108 (2017).
Pyonteck, S. M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013).
Quail, D. F. et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 352, aad3018 (2016). References 64 and 65 describe CSF-1R, an anti-inflammatory target in glioma, and demonstrate that tumour microenvironment-mediated resistance can be overcome by combination with IGF-1R or PI3K inhibitors.
Rommel, C. Taking PI3Kdelta and PI3Kgamma one step ahead: dual active PI3Kdelta/gamma inhibitors for the treatment of immune-mediated inflammatory diseases. Curr. Top. Microbiol. Immunol. 346, 279–299 (2010).
Furman, R. R. et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 370, 997–1007 (2014).
Ali, K. et al. Inactivation of PI(3)K p110delta breaks regulatory T-cell-mediated immune tolerance to cancer. Nature 510, 407–411 (2014).
Ferguson, F. M. et al. Discovery of a Series of 5,11-Dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-ones as Selective PI3K-delta/gamma Inhibitors. ACS Med. Chem. Lett. 7, 908–912 (2016).
O'Brien, S. et al. Duvelisib (IPI-145), a PI3K-δ, γ inhibitor, is clinically active in patients with relapsed/refractory chronic lymphocytic leukemia. Blood 124, 3334–3334 (2014).
Horwitz, S. M. et al. Activity of the PI3K-delta, gamma inhibitor duvelisib in a phase I trial and preclinical models of T-cell lymphoma. Blood https://doi.org/10.1182/blood-2017-08-802470 (2017).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Phan, G. Q. et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc. Natl Acad. Sci. USA 100, 8372–8377 (2003).
Gong, Y. & Pao, W. EGFR mutant lung cancer. Curr. Top. Microbiol. Immunol. 355, 59–81 (2012).
Barouch-Bentov, R. & Sauer, K. Mechanisms of drug-resistance in kinases. Expert Opin. Investig. Drugs 20, 153–208 (2011).
Vanneman, M. & Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 12, 237–251 (2012).
Gainor, J. F. et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin. Cancer Res. 22, 4585–4593 (2016).
Shin, D. S. & Ribas, A. The evolution of checkpoint blockade as a cancer therapy: what's here, what's next? Curr. Opin. Immunol. 33, 23–35 (2015).
Mahoney, K. M., Rennert, P. D. & Freeman, G. J. Combination cancer immunotherapy and new immuno-modulatory targets. Nat. Rev. Drug Discov. 14, 561–584 (2015).
Tchekmedyian, N. et al. Propelling immunotherapy combinations into the clinic. Oncology 29, 990–1002 (2015).
Kaneda, M. M. et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016).
Schmid, M. C. et al. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3kgamma, a single convergent point promoting tumor inflammation and progression. Cancer Cell 19, 715–727 (2011).
Diaz-Montero, C. M., Finke, J. & Montero, A. J. Myeloid-derived suppressor cells in cancer: therapeutic, predictive, and prognostic implications. Semin. Oncol. 41, 174–184 (2014).
Gebhardt, C. et al. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with ipilimumab. Clin. Cancer Res. 21, 5453–5459 (2015).
De Henau, O. et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539, 443–447 (2016).
Evans, C. A. et al. Discovery of a selective phosphoinositide-3-kinase (PI3K)-gamma inhibitor (IPI-549) as an immuno-oncology clinical candidate. ACS Med. Chem. Lett. 7, 862–867 (2016).
Ebert, P. J. et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 44, 609–621 (2016).
Serrels, A. et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell 163, 160–173 (2015).
Stokes, J. B. et al. Inhibition of focal adhesion kinase by PF-562,271 inhibits the growth and metastasis of pancreatic cancer concomitant with altering the tumor microenvironment. Mol. Cancer Ther. 10, 2135–2145 (2011).
Jiang, H. et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 22, 851–860 (2016).
Huang, S. H., Li, Y., Zhang, J., Rong, J. & Ye, S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 31, 330–335 (2013).
Ahn, M. J., Sun, J. M., Lee, S. H., Ahn, J. S. & Park, K. EGFR TKI combination with immunotherapy in non-small cell lung cancer. Expert Opin. Drug Safety 16, 465–469 (2017).
Akbay, E. A. et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 3, 1355–1363 (2013). This paper describes the immunosuppressive functions of oncogenic EGFR and demonstrates synergy between EGFR inhibitors and T cell checkpoint inhibitors.
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Deng, J. et al. CDK4/6 inhibition augments anti-tumor immunity by enhancing T cell activation. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-17-0915 (2017).
Vladimer, G. I. et al. Global survey of the immuno-modulatory potential of common drugs. Nat. Chem. Biol. 13, 681–690 (2017). This paper characterizes the 1,024 FDA-approved drugs in a cell–cell interaction assay and demonstrates that up to 10% have effects on the immune system.
Gould, S. E., Junttila, M. R. & de Sauvage, F. J. Translational value of mouse models in oncology drug development. Nat. Med. 21, 431–439 (2015).
Fleischmann, R. et al. Placebo-controlled trial of tofacitinib monotherapy in rheumatoid arthritis. N. Engl. J. Med. 367, 495–507 (2012).
Wollenhaupt, J. et al. Safety and efficacy of tofacitinib, an oral janus kinase inhibitor, for the treatment of rheumatoid arthritis in open-label, longterm extension studies. J. Rheumatol. 41, 837–852 (2014).
Kawasaki, T. & Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 5, 461 (2014).
Hernandez-Florez, D. & Valor, L. Protein-kinase inhibitors: a new treatment pathway for autoimmune and inflammatory diseases? Rheumatol. Clin. 12, 91–99 (2016).
Patterson, H., Nibbs, R., McInnes, I. & Siebert, S. Protein kinase inhibitors in the treatment of inflammatory and autoimmune diseases. Clin. Exp. Immunol. 176, 1–10 (2014).
Villarino, A. V., Kanno, Y. & O'Shea, J. J. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 18, 374–384 (2017).
Schwartz, D. M., Bonelli, M., Gadina, M. & O'Shea, J. J. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat. Rev. Rheumatol. 12, 25–36 (2016). This Review summarizes the progress in the field of JAK inhibitors for inflammatory disease.
Kontzias, A., Laurence, A., Gadina, M. & O'Shea, J. J. Kinase inhibitors in the treatment of immune-mediated disease. F1000 Med. Rep. 4, 5 (2012).
Cohen, P. Targeting protein kinases for the development of anti-inflammatory drugs. Curr. Opin. Cell Biol. 21, 317–324 (2009).
Weinblatt, M. E. et al. An oral spleen tyrosine kinase (Syk) inhibitor for rheumatoid arthritis. N. Engl. J. Med. 363, 1303–1312 (2010).
Bahjat, F. R. et al. An orally bioavailable spleen tyrosine kinase inhibitor delays disease progression and prolongs survival in murine lupus. Arthritis Rheum. 58, 1433–1444 (2008).
Bajpai, M. Fostamatinib, a Syk inhibitor prodrug for the treatment of inflammatory diseases. IDrugs 12, 174–185 (2009).
Gharwan, H. & Groninger, H. Kinase inhibitors and monoclonal antibodies in oncology: clinical implications. Nat. Rev. Clin. Oncol. 13, 209–227 (2016).
Rankin, A. L. et al. Selective inhibition of BTK prevents murine lupus and antibody-mediated glomerulo-nephritis. J. Immunol. 191, 4540–4550 (2013).
Evans, E. K. et al. Inhibition of Btk with CC-292 provides early pharmacodynamic assessment of activity in mice and humans. J. Pharmacol. Exp. Ther. 346, 219–228 (2013).
Crofford, L. J., Nyhoff, L. E., Sheehan, J. H. & Kendall, P. L. The role of Bruton's tyrosine kinase in autoimmunity and implications for therapy. Expert Rev. Clin. Immunol. 12, 763–773 (2016).
Wu, H. et al. Irreversible inhibition of BTK kinase by a novel highly selective inhibitor CHMFL-BTK-11 suppresses inflammatory response in rheumatoid arthritis model. Sci. Rep. 7, 466 (2017).
Burger, J. A. Bruton's tyrosine kinase (BTK) inhibitors in clinical trials. Curr. Hematol. Malig. Rep. 9, 44–49 (2014).
Dudhgaonkar, S. et al. Selective IRAK4 inhibition attenuates disease in murine lupus models and demonstrates steroid sparing activity. J. Immunol. 198, 1308–1319 (2017).
McElroy, W. T. et al. Potent and selective amidopyrazole inhibitors of IRAK4. ACS Med. Chem. Lett. 6, 677–682 (2015).
Rhyasen, G. W. et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell 24, 90–104 (2013).
Fiore, M., Forli, S. & Manetti, F. Targeting mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2, MK2): medicinal chemistry efforts to lead small molecule inhibitors to clinical trials. J. Med. Chem. 59, 3609–3634 (2016).
Rommel, C., Camps, M. & Ji, H. PI3K delta and PI3K gamma: partners in crime in inflammation in rheumatoid arthritis and beyond? Nat. Rev. Immunol. 7, 191–201 (2007).
Boyle, D. L., Kim, H. R., Topolewski, K., Bartok, B. & Firestein, G. S. Novel phosphoinositide 3-kinase delta, gamma inhibitor: potent anti-inflammatory effects and joint protection in models of rheumatoid arthritis. J. Pharmacol. Exp. Ther. 348, 271–280 (2014).
Winkler, D. G. et al. PI3K-delta and PI3K-gamma inhibition by IPI-145 abrogates immune responses and suppresses activity in autoimmune and inflammatory disease models. Chem. Biol. 20, 1364–1374 (2013).
Olbrich, P. et al. Activated PI3Kdelta syndrome type 2: two patients, a novel mutation, and review of the literature. Pediatr. Allergy Immunol. 27, 640–644 (2016).
Hoegenauer, K. et al. Discovery of CDZ173 (leniolisib), representing a structurally novel class of PI3K delta-selective inhibitors. ACS Med. Chem. Lett. 8, 975–980 (2017).
Rao, V. K. et al. Effective “activated PI3Kdelta syndrome”-targeted therapy with the PI3Kdelta inhibitor leniolisib. Blood 130, 2307–2316 (2017).
Borgel, D. et al. Elevated growth-arrest-specific protein 6 plasma levels in patients with severe sepsis. Crit. Care Med. 34, 219–222 (2006).
Broad, A., Jones, D. E. & Kirby, J. A. Toll-like receptor (TLR) response tolerance: a key physiological “damage limitation” effect and an important potential opportunity for therapy. Curr. Med. Chem. 13, 2487–2502 (2006).
Corbett, A. et al. Drug repositioning for Alzheimer's disease. Nat. Rev. Drug Discov. 11, 833–846 (2012).
McGonigle, P. Animal models of CNS disorders. Biochem. Pharmacol. 87, 140–149 (2014).
Horvath, P. et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 15, 751–769 (2016).
Ghosh, R. et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548 (2014). This paper validates IRE1α kinase inhibition in models of retinitis pigmentosa and diabetes.
Meredith, E. L. et al. Discovery of oral VEGFR-2 inhibitors with prolonged ocular retention that are efficacious in models of wet age-related macular degeneration. J. Med. Chem. 58, 9273–9286 (2015). This paper describes a successful preclinical study of a small-molecule VEGFR2 inhibitor for treatment of wet AMD.
Credle, J. J., Finer-Moore, J. S., Papa, F. R., Stroud, R. M. & Walter, P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 102, 18773–18784 (2005).
Zhou, J. et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl Acad. Sci. USA 103, 14343–14348 (2006).
Calfon, M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002).
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001).
Han, D. et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575 (2009).
Shore, G. C., Papa, F. R. & Oakes, S. A. Signaling cell death from the endoplasmic reticulum stress response. Curr. Opin. Cell Biol. 23, 143–149 (2011).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Moreno, J. A. et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl Med. 5, 206ra138 (2013).
Radford, H., Moreno, J. A., Verity, N., Halliday, M. & Mallucci, G. R. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 130, 633–642 (2015).
Axten, J. M. et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 55, 7193–7207 (2012).
Larhammar, M. et al. Dual leucine zipper kinase-dependent PERK activation contributes to neuronal degeneration following insult. eLife 6, e20725 (2017).
Patel, S. et al. Scaffold-hopping and structure-based discovery of potent, selective, and brain penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine inhibitors of dual leucine zipper kinase (DLK. MAP3K12). J. Med. Chem. 58, 8182–8199 (2015).
Patel, S. et al. Discovery of dual leucine zipper kinase (DLK, MAP3K12) inhibitors with activity in neurodegeneration models. J. Med. Chem. 58, 401–418 (2015).
Ambati, J. & Fowler, B. J. Mechanisms of age-related macular degeneration. Neuron 75, 26–39 (2012).
Ishikawa, M., Jin, D., Sawada, Y., Abe, S. & Yoshitomi, T. Future therapies of wet age-related macular degeneration. J. Ophthalmol. 2015, 138070 (2015).
Appelmann, I., Liersch, R., Kessler, T., Mesters, R. M. & Berdel, W. E. Angiogenesis inhibition in cancer therapy: platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) and their receptors: biological functions and role in malignancy. Recent results in cancer research. Recent Results Cancer Res. 180, 51–81 (2010).
Diago, T. Jr. et al., Ranibizumab combined with low-dose sorafenib for exudative age-related macular degeneration. Mayo Clin. Proc. 83, 231–234 (2008).
Slakter, J. S. et al. Phase I/II study of oral pazopanib, a receptor tyrosine kinase inhibitor, in neovascular age related macular degeneration. Invest. Ophthalmol. Visual Sci. 53, 2038 (2012).
Liang, C., Brown, D., Chaudhry, N., Elman, M. & Heier, J. Rationale for treating wet AMD in human using an oral pill consisting of a VEGFR/PDGFR inhibitor X-82. Invest. Ophthalmol. Visual Sci. 54, 3272–3272 (2013).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02535286 (2017).
Mainolfi, N., Karki, R., Liu, F. & Anderson, K. Evolution of a new class of VEGFR-2 inhibitors from scaffold morphing and redesign. ACS Med. Chem. Lett. 7, 363–367 (2016).
Artac, R. A. et al. Neutralization of vascular endothelial growth factor antiangiogenic isoforms is more effective than treatment with proangiogenic isoforms in stimulating vascular development and follicle progression in the perinatal rat ovary. Biol. Reprod. 81, 978–988 (2009).
Batson, J. et al. Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease. ACS Chem. Biol. 12, 825–832 (2017).
Scott, J. D. et al. Discovery of a 3-(4-Pyrimidinyl) indazole (MLi-2), an orally available and selective leucine-rich repeat kinase 2 (LRRK2) inhibitor that reduces brain kinase activity. J. Med. Chem. 60, 2983–2992 (2017).
Heffron, T. P. et al. Discovery of clinical development candidate GDC-0084, a brain penetrant inhibitor of PI3K and mTOR. ACS Med. Chem. Lett. 7, 351–356 (2016).
Baltussen, L., Rosianu, F. & Ultanir, S. Kinases in synaptic development and neurological diseases. Prog. Neuropsychopharmacol. Biol. Psychiatry https://doi.org/10.1016/j.pnpbp.2017.12.006 (2017).
Switon, K., Kotulska, K., Janusz-Kaminska, A., Zmorzynska, J. & Jaworski, J. Molecular neurobiology of mTOR. Neuroscience 341, 112–153 (2017).
Chan, S. L. & Tan, E. K. Targeting LRRK2 in Parkinson's disease: an update on recent developments. Expert Opin. Ther. Targets 21, 601–610 (2017).
Lucet, I. S., Tobin, A., Drewry, D., Wilks, A. F. & Doerig, C. Plasmodium kinases as targets for new-generation antimalarials. Future Med. Chem. 4, 2295–2310 (2012).
McNamara, C. W. et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504, 248–253 (2013). This thorough study validates PfPI(4)K as an antimalarial target.
Crowther, G. J. et al. Biochemical screening of five protein kinases from Plasmodium falciparum against 14,000 cell-active compounds. PLoS ONE 11, e0149996 (2016).
Derbyshire, E. R. et al. Chemical interrogation of the malaria kinome. Chembiochem 15, 1920–1930 (2014).
Carr, J. M., Mahalingam, S., Bonder, C. S. & Pitson, S. M. Sphingosine kinase 1 in viral infections. Rev. Med. Virol. 23, 73–84 (2013).
Eisa-Beygi, S. & Wen, X. Y. Could pharmacological curtailment of the RhoA/Rho-kinase pathway reverse the endothelial barrier dysfunction associated with Ebola virus infection? Antiviral Res. 114, 53–56 (2015).
Clark, M. J. et al. GNF-2 inhibits dengue virus by targeting Abl kinases and the viral E protein. Cell Chem. Biol. 23, 443–452 (2016).
Schreiber, M., Res, I. & Matter, A. Protein kinases as antibacterial targets. Curr. Opin. Cell Biol. 21, 325–330 (2009).
Gordon, S., Simithy, J., Goodwin, D. C. & Calderon, A. I. Selective Mycobacterium tuberculosis shikimate kinase inhibitors as potential antibacterials. Persp. Med. Chem. 7, 9–20 (2015).
Prisic, S. & Husson, R. N. Mycobacterium tuberculosis serine/threonine protein kinases. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.MGM2-0006-2013 (2014).
Wang, T. et al. Mtb PKNA/PKNB dual inhibition provides selectivity advantages for inhibitor design to minimize host kinase interactions. ACS Med. Chem. Lett. 8, 1224–1229 (2017).
Saleh, D. & Degterev, A. Emerging roles for RIPK1 and RIPK3 in Pathogen-induced cell death and host immunity. Curr. Top. Microbiol. Immunol. 403, 37–75 (2017).
Volpe, G., Panuzzo, C., Ulisciani, S. & Cilloni, D. Imatinib resistance in CML. Cancer Lett. 274, 1–9 (2009).
Niederst, M. J. & Engelman, J. A. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci. Signal. 6, re6 (2013).
Smyth, L. A. & Collins, I. Measuring and interpreting the selectivity of protein kinase inhibitors. J. Chem. Biol. 2, 131–151 (2009).
Miduturu, C. V. et al. High-Throughput Kinase Profiling: A More Efficient Approach towards the Discovery of New Kinase Inhibitors. Chem. Biol. 18, 868–879 (2011).
Cully, M. Rational drug design: Tuning kinase inhibitor residence time. Nat. Rev. Drug Discov. 14, 457 (2015).
Zhang, J., Yang, P. L. & Gray, N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Sang, J. et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov. 3, 430–443 (2013). This paper describes EMAP4–ALK as a highly sensitive client of HSP90, which is preferentially degraded upon HSP90 inhibition.
Katayama, R. et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc. Natl Acad. Sci. USA 108, 7535–7540 (2011).
Richards, M. W. et al. Crystal structure of EML1 reveals the basis for Hsp90 dependence of oncogenic EML4-ALK by disruption of an atypical beta-propeller domain. Proc. Natl Acad. Sci. USA 111, 5195–5200 (2014).
Workman, P. & van Montfort, R. EML4-ALK fusions: propelling cancer but creating exploitable chaperone dependence. Cancer Discov. 4, 642–645 (2014).
Chen, Z. et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 70, 9827–9836 (2010).
Wang, M. et al. Development of heat shock protein (Hsp90) inhibitors to combat resistance to tyrosine kinase inhibitors through Hsp90-kinase interactions. J. Med. Chem. 59, 5563–5586 (2016).
Kronke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523, 183–188 (2015). This paper uncovers the mechanism of action of lenalidomide in myelodisplastic syndrome.
Petzold, G., Fischer, E. S. & Thoma, N. H. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 532, 127–130 (2016). This paper characterizes the structural basis for small-molecule-induced CRBN–CKI-α dimerization, providing rationale for development of molecules hijacking this pathway for induced degradation approaches.
Beke, L. et al. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci. Rep. 35, e00267 (2015).
Kii, I. et al. Selective inhibition of the kinase DYRK1A by targeting its folding process. Nat. Commun. 7, 11391 (2016).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Kritzer, J. New Frontiers in Chemical Biology: Enabling Drug Discovery. Edited by Mark E. Bunnage. ChemMedChem 6, 1747–1748 (2011).
Buckley, D. L. & Crews, C. M. Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system. Angew. Chem. Int. Ed. 53, 2312–2330 (2014).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017). This paper provides an extensive and up-to-date review of the PROTAC field.
Toure, M. & Crews, C. M. Small-molecule PROTACS: new approaches to protein degradation. Angew. Chem. Int. Ed. 55, 1966–1973 (2016).
Henning, R. K. et al. Degradation of Akt using protein-catalyzed capture agents. J. Pept. Sci. 22, 196–200 (2016).
Lai, A. C. et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. 55, 807–810 (2016).
Hines, J., Gough, J. D., Corson, T. W. & Crews, C. M. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc. Natl Acad. Sci. USA 110, 8942–8947 (2013).
Crew, A. P. et al. Identification and characterization of Von Hippel-Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J. Med. Chem. 61, 583–598 (2018).
Robb, C. M. et al. Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC). Chem. Commun. 53, 7577–7580 (2017).
Olson, C. M. et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 14, 163–170 (2018).
Xie, T. et al. Pharmacological targeting of the pseudokinase Her3. Nat. Chem. Biol. 10, 1006–1012 (2014).
Chan, K. H., Zengerle, M., Testa, A. & Ciulli, A. Impact of target warhead and linkage vector on inducing protein degradation: comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J. Med. Chem. 61, 504–513 (2018).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Matyskiela, M. E. et al. Cereblon modulator (CC-220) with improved degradation of Ikaros and Aiolos. J. Med. Chem. 61, 535–542 (2018).
Hansen, J. D. et al. Protein degradation via CRL4CRBN ubiquitin ligase: discovery and structure-activity relationships of novel glutarimide analogs that promote degradation of Aiolos and/or GSPT1. J. Med. Chem. 61, 492–503 (2018).
Huang, H. T. et al. Chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 25, 88–99.e6 (2018).
Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78–87.e5 (2018).
Yang, C. et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 36, 2255–2264 (2017).
Herrera-Abreu, M. T. et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res. 76, 2301–2313 (2016).
Taavi, K. et al. An oral androgen receptor PROTAC degrader for prostate cancer [abstract]. J. Clin. Oncol. 35 (Suppl.), 273 (2017).
Leestemaker, Y. et al. Proteasome activation by small molecules. Cell Chem. Biol. 24, 725–736.e7 (2017).
Liu, Q. et al. Developing irreversible inhibitors of the protein kinase cysteinome. Chem. Biol. 20, 146–159 (2013).
Chaikuad, A., Koch, P., Laufer, S. & Knapp, S. Targeting the protein kinases cysteinome. Angew. Chem. Int. Ed. https://doi.org/10.1002/anie.201707875 (2017).
Fischer, P. M. Approved and experimental small-molecule oncology kinase inhibitor drugs: a mid-2016 overview. Med. Res. Rev. 37, 314–367 (2017).
Strelow, J. M. A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov. 22, 3–20 (2017).
Copeland, R. A. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods Biochem. Anal. 46, 1–265 (2005).
Mohutsky, M. & Hall, S. D. Irreversible enzyme inhibition kinetics and drug-drug interactions. Methods Mol. Biol. 1113, 57–91 (2014).
Schwartz, P. A. et al. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl Acad. Sci. USA 111, 173–178 (2014).
Zaro, B. W., Whitby, L. R., Lum, K. M. & Cravatt, B. F. Metabolically Labile Fumarate Esters Impart Kinetic Selectivity to Irreversible Inhibitors. J. Am. Chem. Soc. 138, 15841–15844 (2016). This paper describes a novel cysteine targeting warhead with improved on-target selectivity.
Serafimova, I. M. et al. Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol. 8, 471–476 (2012).
Krishnan, S. et al. Design of reversible, cysteine-targeted Michael acceptors guided by kinetic and computational analysis. J. Am. Chem. Soc. 136, 12624–12630 (2014).
Bradshaw, J. M. et al. Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat. Chem. Biol. 11, 525–531 (2015). This paper reports a method for rational design of drug residence times via reversible covalent interactions.
Forster, M. et al. Selective JAK3 inhibitors with a covalent reversible binding mode targeting a new induced fit binding pocket. Cell Chem. Biol. 23, 1335–1340 (2016).
London, N. et al. Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol. 10, 1066–1072 (2014).
Miller, R. M., Paavilainen, V. O., Krishnan, S., Serafimova, I. M. & Taunton, J. Electrophilic fragment-based design of reversible covalent kinase inhibitors. J. Am. Chem. Soc. 135, 5298–5301 (2013).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016). This paper uses reactive fragments to enumerate the targetable cysteine residues in the proteome.
Anscombe, E. et al. Identification and characterization of an irreversible inhibitor of CDK2. Chem. Biol. 22, 1159–1164 (2015).
Lin, S. et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602 (2017).
Dalton, S. E. et al. Selectively targeting the kinome-conserved lysine of PI3Kdelta as a general approach to covalent kinase inhibition. J. Am. Chem. Soc. 140, 932–939 (2018).
Healy, D. G. et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 7, 583–590 (2008).
Greggio, E. & Cookson, M. R. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro 1, e00002 (2009).
Kumar, A. & Cookson, M. R. Role of LRRK2 kinase dysfunction in Parkinson disease. Expert Rev. Mol. Med. 13, e20 (2011).
West, A. B. Achieving neuroprotection with LRRK2 kinase inhibitors in Parkinson disease. Exp. Neurol. 298, 236–245 (2017).
Lee, K. L. et al. Discovery of clinical candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoli ne-6-carboxamide (PF-06650833), a potent, selective inhibitor of interleukin-1 receptor associated kinase 4 (IRAK4), by fragment-based drug design. J. Med. Chem. 60, 5521–5542 (2017).
Gomez, N., Erazo, T. & Lizcano, J. M. ERK5 and cell proliferation: nuclear localization is what matters. Front. Cell Dev. Biol. 4, 105 (2016).
Hoang, V. T. et al. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 392, 51–59 (2017).
Lin, E. C. et al. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl Acad. Sci. USA 113, 11865–11870 (2016).
Deng, X. et al. Discovery of a benzo[e]pyrimido-[5,4-b][1,4]diazepin-6(11H)-one as a potent and selective inhibitor of big MAP kinase 1. ACS Med. Chem. Lett. 2, 195–200 (2011).
Deng, X. et al. Structural determinants for ERK5 (MAPK7) and leucine rich repeat kinase 2 activities of benzo[e]pyrimido-[5,4-b]diazepine-6(11H)-ones. Eur. J. Med. Chem. 70, 758–767 (2013).
Elkins, J. M. et al. X-Ray crystal structure of ERK5 (MAPK7) in complex with a specific inhibitor. J. Med. Chem. 56, 4413–4421 (2013).
Berger, S. B. et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480 (2014).
Harris, P. A. et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J. Med. Chem. 59, 2163–2178 (2016).
Harris, P. A. et al. Discovery of a first-in-class receptor interacting protein 1 (RIP1) kinase specific clinical candidate (GSK2982772) for the treatment of inflammatory diseases. J. Med. Chem. 60, 1247–1261 (2017).
Goldstein, D. M., Gray, N. S. & Zarrinkar, P. P. High-throughput kinase profiling as a platform for drug discovery. Nat. Rev. Drug Discov. 7, 391–397 (2008).
Nomura, D. K., Dix, M. M. & Cravatt, B. F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 10, 630–638 (2010).
Patricelli, M. P. et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 18, 699–710 (2011).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Medard, G. et al. Optimized chemical proteomics assay for kinase inhibitor profiling. J. Proteome Res. 14, 1574–1586 (2015).
Golkowski, M. et al. Kinobead and single-shot LC-MS profiling identifies selective PKD inhibitors. J. Proteome Res. 16, 1216–1227 (2017).
Vasta, J. D. et al. Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem. Biol. https://doi.org/10.1016/j.chembiol.2017.10.010 (2017). This paper describes a commercially available, in cell competition assay to discern kinase inhibitor selectivity in a physiological context.
Munoz, L. Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 16, 424–440 (2017). This article is an overview of unexpected off-target activities that have been discovered in kinase inhibitors.
Ciceri, P. et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat. Chem. Biol. 10, 305–312 (2014).
Ember, S. W. et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem. Biol. 9, 1160–1171 (2014).
Rimassa, L., Bruix, J., Broggini, M. & Santoro, A. Tivantinib (ARQ197) displays cytotoxic activity that is independent of its ability to bind MET — letter. Clin. Cancer Res. 19, 4290 (2013).
Katayama, R. et al. Cytotoxic activity of tivantinib (ARQ 197) is not due solely to c-MET inhibition. Cancer Res. 73, 3087–3096 (2013).
Cheong, J. K. et al. IC261 induces cell cycle arrest and apoptosis of human cancer cells via CK1delta/varepsilon and Wnt/beta-catenin independent inhibition of mitotic spindle formation. Oncogene 30, 2558–2569 (2011).
Lanning, B. R. et al. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat. Chem. Biol. 10, 760–767 (2014). This paper describes pull-down proteomics inhibitor profiling methods for assessing the proteome-wide selectivity of covalent inhibitors.
Niphakis, M. J. & Cravatt, B. F. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 83, 341–377 (2014).
Yang, P. & Liu, K. Activity-based protein profiling: recent advances in probe development and applications. Chembiochem 16, 712–724 (2015).
Wang, K. et al. Chemistry-based functional proteomics for drug target deconvolution. Expert Rev. Proteom. 9, 293–310 (2012).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Klaeger, S. et al. Chemical proteomics reveals ferrochelatase as a common off-target of kinase inhibitors. ACS Chem. Biol. 11, 1245–1254 (2016).
Reinhard, F. B. et al. Thermal proteome profiling monitors ligand interactions with cellular membrane proteins. Nat. Methods 12, 1129–1131 (2015).
Kooistra, A. J. et al. KLIFS: a structural kinase-ligand interaction database. Nucleic Acids Res. 44, D365–D371 (2016).
Isberg, V. et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 44, D356–D364 (2016).
McGuire, R. et al. 3D-e-Chem-VM: structural cheminformatics research infrastructure in a freely available virtual machine. J. Chem. Inform. Model. 57, 115–121 (2017).
Lin, X. et al. Life beyond kinases: structure-based discovery of sorafenib as nanomolar antagonist of 5-HT receptors. J. Med. Chem. 55, 5749–5759 (2012).
Merget, B., Turk, S., Eid, S., Rippmann, F. & Fulle, S. Profiling prediction of kinase inhibitors: toward the virtual assay. J. Med. Chem. 60, 474–485 (2017).
[No authors listed.] Kinase Assay Tools. Reaction Biology Corp. http://www.reactionbiology.com/webapps/site/KinaseDetail.aspx (2016).
Drewry, D. H. et al. Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS ONE 12, e0181585 (2017).
Klaeger, S. et al. The target landscape of clinical kinase drugs. Science 358, eaan4368 (2017).
Leelananda, S. P. & Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 12, 2694–2718 (2016).
Li, Z. & Lazaridis, T. Thermodynamic contributions of the ordered water molecule in HIV-1 protease. J. Am. Chem. Soc. 125, 6636–6637 (2003).
Huggins, D. J., Quantifying the entropy of binding for water molecules in protein cavities by computing correlations. Biophys. J. 108, 928–936 (2015).
Li, Z. & Lazaridis, T. Computing the thermodynamic contributions of interfacial water. Methods Mol. Biol. 819, 393–404 (2012).
Robinson, D. D., Sherman, W. & Farid, R. Understanding kinase selectivity through energetic analysis of binding site waters. ChemMedChem 5, 618–627 (2010).
Abel, R., Young, T., Farid, R., Berne, B. J. & Friesner, R. A. Role of the active-site solvent in the thermodynamics of factor Xa ligand binding. J. Am. Chem. Soc. 130, 2817–2831 (2008).
Young, T., Abel, R., Kim, B., Berne, B. J. & Friesner, R. A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc. Natl Acad. Sci. USA 104, 808–813 (2007).
Kitamura, K. et al. Binding free-energy calculation is a powerful tool for drug optimization: calculation and measurement of binding free energy for 7-azaindole derivatives to glycogen synthase kinase-3beta. J. Chem. Inform. Model. 54, 1653–1660 (2014). This study is an elegant example of the accuracy of FEP calculations in ranking kinase inhibitors.
Lin, Y. L., Meng, Y., Jiang, W. & Roux, B. Explaining why Gleevec is a specific and potent inhibitor of Abl kinase. Proc. Natl Acad. Sci. USA 110, 1664–1669 (2013).
Araki, M. et al. The effect of conformational flexibility on binding free energy estimation between kinases and their inhibitors. J. Chem. Inform. Model. 56, 2445–2456 (2016).
Ruiz-Carmona, S. et al. Dynamic undocking and the quasi-bound state as tools for drug discovery. Nat. Chem. 9, 201–206 (2017). This paper describes a new method for virtual screening and ranking ligands on the basis of calculations of nonthermodynamic properties.
Takeuchi, K. & Ito, F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 34, 1774–1780 (2011).
Mushtaq, G. et al. Neuroprotective mechanisms mediated by CDK5 inhibition. Curr. Pharm. Design 22, 527–534 (2016).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02784106 (2017).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT01882803 (2017).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02100852 (2017).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT01674569 (2017).
Narayanan, A. & Jones, L. H. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 6, 2650–2659 (2015).
Morales-Sanfrutos, J. et al. Vinyl sulfone: a versatile function for simple bioconjugation and immobilization. Org. Biomol. Chem. 8, 667–675 (2010).
Garcia, F. J. & Carroll, K. S. Redox-based probes as tools to monitor oxidized protein tyrosine phosphatases in living cells. Eur. J. Med. Chem. 88, 28–33 (2014).
Leonard, S. E., Garcia, F. J., Goodsell, D. S. & Carroll, K. S. Redox-based probes for protein tyrosine phosphatases. Angew. Chem. Int. Ed. 50, 4423–4427 (2011).
Wani, R. et al. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc. Natl Acad. Sci. USA 108, 10550–10555 (2011).
Acknowledgements
The authors thank B.J. Pinch, D.A. Scott, N.P. Kwiatkowski and C.M. Olson for proofreading and comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
N.S.G. is a scientific founder and equity holder in C4, Petra, Syros and Gatekeeper Pharmaceuticals.
Glossary
- Free-energy perturbation
-
(FEP). A method for computing free-energy differences from molecular dynamics simulations on the basis of statistical mechanics.
- Super-enhancers
-
(SEs). Large clusters of transcriptional enhancers, which normally drive expression of genes that define cell identity. Tumour cells acquire SEs at oncogenes and at genes associated with the acquisition of the hallmarks of cancer.
- Tumour microenvironment
-
The cellular environment in which the tumour exists, which is composed of surrounding blood vessels, immune cells, fibroblasts, inflammatory cells, lymphocytes, signalling molecules and the extracellular matrix. The tumour microenvironment and the tumour are constantly interacting; thus, the tumour microenvironment affects tumour development and progression.
- T cell checkpoint inhibitors
-
Antibody-based therapies that block inhibitory pathways that regulate the adaptive immune response. Currently approved T cell checkpoint inhibitors target programmed cell death protein 1 (PD1), PDL1 (PD1 ligand 1) and cytotoxic T lymphocyte-associated protein 4 (CTLA4).
- Natural killer cells
-
Lymphocytes able to bind to certain tumour cells and virus-infected cells and kill them by the injection of granzymes. Unlike T cells, natural killer cells can recognize stressed cells in the absence of antibodies or major histocompatibility complex (MHC) expression, and elicit rapid immune responses.
- PROTACs
-
Proteolysis-targeting chimaeras (PROTACs) are two-headed molecules capable of removing unwanted proteins by inducing selective intracellular proteolysis through induction of their ubiquitylation.
- Covalent kinase inhibitors
-
Kinase inhibitors that contain a weakly reactive electrophile. Upon a reversible binding interaction, the electrophilic warhead is brought into close proximity with a nucleophilic residue in the kinase, often cysteine, which subsequently reacts to form a covalently bonded complex.
- DNA-encoded library
-
Contains small molecules conjugated to short DNA fragments that serve as identification barcodes. The technique enables the mass interrogation of chemical libraries via affinity selection, typically on an immobilized protein target, followed by amplification of the binder's 'barcodes' via PCR and compound identification by DNA sequencing. The advantage of this technique is its screening efficiency.
- Alchemical pathways
-
Simulated pathways from one physical state to another, that proceed via a series of non-physical intermediates.
Rights and permissions
About this article
Cite this article
Ferguson, F., Gray, N. Kinase inhibitors: the road ahead. Nat Rev Drug Discov 17, 353–377 (2018). https://doi.org/10.1038/nrd.2018.21
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2018.21
This article is cited by
-
CircHAS2 activates CCNE2 to promote cell proliferation and sensitizes the response of colorectal cancer to anlotinib
Molecular Cancer (2024)
-
The protein phosphatase-2A subunit PR130 is involved in the formation of cytotoxic protein aggregates in pancreatic ductal adenocarcinoma cells
Cell Communication and Signaling (2024)
-
Discovery of a nitroaromatic nannocystin with potent in vivo anticancer activity against colorectal cancer by targeting AKT1
Acta Pharmacologica Sinica (2024)
-
Iterative machine learning-based chemical similarity search to identify novel chemical inhibitors
Journal of Cheminformatics (2023)
-
The ULK3 kinase is a determinant of keratinocyte self-renewal and tumorigenesis targeting the arginine methylome
Nature Communications (2023)