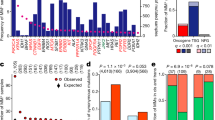
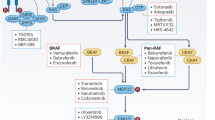
Abstract
Despite the development of drugs inhibiting the oncogenic proteins that cancer cells are dependent on, attempts to match targeted therapies to the genetic makeup of individual tumors is proving more difficult than expected. Until now, the paradigm has been a binary correlation between a mutated cancer gene and response to a given therapy. However, recent evidence indicates that different genetic alterations, such as mutations in different codons of a cancer gene, might be related to distinct sensitivity to targeted therapies. An example is the divergent effect that individual EGFR, PIK3CA and KRAS mutations might have on response or resistance to tailored drugs. Furthermore, the idea that the presence of a specific mutation translates into sensitivity or resistance to a particular drug is likely too simplistic, since it does not capture the complexity of the signaling pathways in an individual cancer. Only the overall genetic milieu (alterations in upstream and/or parallel pathways) ultimately determines the response of individual tumors to therapy. We have critically analyzed data supporting the genetic, biological and biochemical differences of individual mutations within a single cancer gene. The role of cancer mutations as predictors of sensitivity and resistance to targeted therapies is discussed, together with the implications for the 'personalized' treatment of cancer patients.
Key Points
-
Cancer is—in essence—a genetic disease and oncogenic mutations are legitimate targets for therapy
-
Genetic alterations are responsible for distinct sensitivity to targeted therapies and their detection can be used to direct therapy against definite patient populations
-
Different mutations within an individual gene might result in divergent degrees of response and resistance to therapy
-
The overall molecular makeup (and not the individual mutations) determines the response of a tumor to therapy
-
Alterations in cancer genes can exert different effects on response to therapy depending on the tumor type in which they occur
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tejpar, S. et al. Mutant KRAS and BRAF gene expression profiles in colorectal cancer: Results of the translational study on the PETACC 3-EORTC 40993-SAKK 60–00 trial [abstract]. J. Clin. Oncol. 28 (Suppl.), a3511 (2010).
Wirapati, P. et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 10, R65 (2008).
Blows, F. M. et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 7, e1000279 (2010).
Cetin, K., Ettinger, D. S., Hei, Y. J. & O'Malley, C. D. Survival by histologic subtype in stage IV nonsmall cell lung cancer based on data from the Surveillance, Epidemiology and End Results Program. Clin. Epidemiol. 3, 139–148 (2011).
Roth, J. A. & Carlson, J. J. Prognostic role of ERCC1 in advanced non-small-cell lung cancer: a systematic review and meta-analysis. Clin. Lung Cancer 12, 393–401 (2011).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011).
Welcome Trust Sanger Institute. Scientific Resources [online], (2011).
Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007).
Kinzler, K. W. & Vogelstein, B. Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996).
Torkamani, A., Verkhivker, G. & Schork, N. J. Cancer driver mutations in protein kinase genes. Cancer Lett. 281, 117–127 (2009).
EMA. European Medicines Agency [online], (2011).
FDA. U.S. Food and Drug Administration [online], (2011).
Druker, B. J. et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 2, 561–566 (1996).
Hirota, S. et al. Gain-of-function mutation at the extracellular domain of KIT in gastrointestinal stromal tumours. J. Pathol. 193, 505–510 (2001).
Heinrich, M. C. et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299, 708–710 (2003).
Antonescu, C. R. et al. Association of KIT exon 9 mutations with nongastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin. Cancer Res. 9, 3329–3337 (2003).
Blanke, C. D. et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J. Clin. Oncol. 26, 620–625 (2008).
Lasota, J. et al. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. A study of 200 cases. Am. J. Pathol. 157, 1091–1095 (2000).
Heinrich, M. C. et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 21, 4342–4349 (2003).
Debiec-Rychter, M. et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur. J. Cancer 42, 1093–1103 (2006).
Slamon, D. J. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244, 707–712 (1989).
Gianni, L. et al. Treatment with trastuzumab for 1 year after adjuvant chemotherapy in patients with HER2-positive early breast cancer: a 4-year follow-up of a randomised controlled trial. Lancet Oncol. 12, 236–244 (2011).
Esteva, F. J. et al. Phase II study of weekly docetaxel and trastuzumab for patients with HER-2-overexpressing metastatic breast cancer. J. Clin. Oncol. 20, 1800–1808 (2002).
Morrow, P. K. et al. Phase I/II study of trastuzumab in combination with everolimus (RAD001) in patients with HER2-overexpressing metastatic breast cancer who progressed on trastuzumab-based therapy. J. Clin. Oncol. 29, 3126–3132 (2011).
Nahta, R., Hung, M. C. & Esteva, F. J. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 64, 2343–2346 (2004).
Seidman, A. D. et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of Cancer and Leukemia Group B protocol 9840. J. Clin. Oncol. 26, 1642–1649 (2008).
Pritchard, K. I. et al. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N. Engl. J. Med. 354, 2103–2111 (2006).
Wolff, A. C. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 25, 118–145 (2007).
Kaufman, P. A. et al. CALGB 150002: Correlation of HER2 and chromosome 17 (ch17) copy number with trastuzumab (T) efficacy in CALGB 9840, paclitaxel (P) with or without T in HER2+ and HER2– metastatic breast cancer (MBC) [abstract]. J. Clin. Oncol. 25 (Suppl.), a1009 (2007).
Perez, E. A. et al. HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. J. Clin. Oncol. 28, 4307–4315 (2010).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Vogel, C. L. et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 20, 719–726 (2002).
Nahta, R., Yu, D., Hung, M. C., Hortobagyi, G. N. & Esteva, F. J. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat. Clin. Pract. Oncol. 3, 269–280 (2006).
Li, J. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997).
Nagata, Y. et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6, 117–127 (2004).
Saal, L. H. et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 65, 2554–2559 (2005).
Berns, K. et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12, 395–402 (2007).
Razis, E. et al. Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res. Treat. 128, 447–456 (2011).
Esteva, F. J. et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am. J. Pathol. 177, 1647–1656 (2010).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Morinaga, R. et al. Association of epidermal growth factor receptor (EGFR) gene mutations with EGFR amplification in advanced non-small cell lung cancer. Cancer Sci. 99, 2455–2460 (2008).
Cappuzzo, F. et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J. Natl Cancer Inst. 97, 643–655 (2005).
Marks, J. L. et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J. Thorac. Oncol. 3, 111–116 (2008).
Cohen, M. H. et al. United States Food and Drug Administration Drug Approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin. Cancer Res. 10, 1212–1218 (2004).
Sordella, R., Bell, D. W., Haber, D. A. & Settleman, J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 305, 1163–1167 (2004).
Pao, W. & Girard, N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 12, 175–180 (2011).
Gazdar, A. F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 (Suppl. 1), S24–S31 (2009).
Greulich, H. et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2, e313 (2005).
Ichihara, S. et al. The impact of epidermal growth factor receptor gene status on gefitinib-treated Japanese patients with non-small-cell lung cancer. Int. J. Cancer 120, 1239–1247 (2007).
Carey, K. D. et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 66, 8163–8171 (2006).
Mulloy, R. et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 67, 2325–2330 (2007).
Jackman, D. M. et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 3908–3914 (2006).
Riely, G. J. et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 839–844 (2006).
Rosell, R. et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 361, 958–967 (2009).
Won, Y. W. et al. Comparison of clinical outcome of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations. J. Clin. Pathol. 64, 947–952 (2011).
Yasuda, H., Kobayashi, S. & Costa, D. B. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. http://dx.doi.org/10.1016/S1470-2045(11)70129-2.
Shigematsu, H. et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl Cancer Inst. 97, 339–346 (2005).
Hsieh, M. H. et al. Complex mutation patterns of epidermal growth factor receptor gene associated with variable responses to gefitinib treatment in patients with non-small cell lung cancer. Lung Cancer 53, 311–322 (2006).
Chou, T. Y. et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin. Cancer Res. 11, 3750–3757 (2005).
Wu, J. Y. et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin. Cancer Res. 14, 4877–4882 (2008).
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).
Tsai, J. et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl Acad. Sci. USA 105, 3041–3046 (2008).
Joseph, E. W. et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl Acad. Sci. USA 107, 14903–14908 (2010).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
Kopetz, S. et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors [abstract]. J. Clin.. Oncol. 28 (Suppl.), a3534 (2010).
Xing, F. et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene http://dx.doi.org/10.1038/onc.2011.250.
Benvenuti, S. et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 67, 2643–2648 (2007).
Karapetis, C. S. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 359, 1757–1765 (2008).
Lievre, A. et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 66, 3992–3995 (2006).
Loupakis, F. et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br. J. Cancer 101, 715–721 (2009).
Linardou, H. et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 9, 962–972 (2008).
Di Nicolantonio, F. et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 26, 5705–5712 (2008).
De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 11, 753–762 (2010).
Gupta, S. et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 129, 957–968 (2007).
Cantley, L. C. & Neel, B. G. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl Acad. Sci. USA 96, 4240–4245 (1999).
Sartore-Bianchi, A. et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 69, 1851–1857 (2009).
Prenen, H. et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin. Cancer Res. 15, 3184–3188 (2009).
Perrone, F. et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann. Oncol. 20, 84–90 (2009).
Zhao, L. & Vogt, P. K. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl Acad. Sci. USA 105, 2652–2657 (2008).
Frattini, M. et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br. J. Cancer 97, 1139–1145 (2007).
Loupakis, F. et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J. Clin. Oncol. 27, 2622–2629 (2009).
Moroni, M. et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 6, 279–286 (2005).
Tejpar, S. et al. Influence of KRAS G13D mutations on outcome in patients with metastatic colorectal cancer (mCRC) treated with first-line chemotherapy with or without cetuximab [abstract]. J. Clin. Oncol. 29 (Suppl.), a3511 (2011).
De Roock, W. et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304, 1812–1820 (2010).
Janakiraman, M. et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 70, 5901–5911 (2010).
Edkins, S. et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol. Ther. 5, 928–932 (2006).
Feig, L. A. & Cooper, G. M. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell Biol. 8, 2472–2478 (1988).
Bos, J. L. Ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 (1989).
Andreyev, H. J., Norman, A. R., Cunningham, D., Oates, J. R. & Clarke, P. A. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J. Natl Cancer Inst. 90, 675–684 (1998).
Almoguera, C. et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53, 549–554 (1988).
Urban, T. et al. Detection of codon 12 K-ras mutations in non-neoplastic mucosa from bronchial carina in patients with lung adenocarcinomas. Br. J. Cancer 82, 412–417 (2000).
Bennecke, M. et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell 18, 135–146 (2011).
Stemke-Hale, K. et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 (2008).
Vasudevan, K. M. et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 16, 21–32 (2009).
Loi, S. et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc. Natl Acad. Sci. USA 107, 10208–10213 (2010).
Chmielecki, J. et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci. Transl. Med. 3, 90ra59 (2011).
Fodde, R., Smits, R. & Clevers, H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1, 55–67 (2001).
Fearnhead, N. S., Britton, M. P. & Bodmer, W. F. The ABC of APC. Hum. Mol. Genet. 10, 721–733 (2001).
Acknowledgements
We wish to thank Federica Di Nicolantonio for providing valuable feedback and Mariangela Russo and Sabrina Arena for critically reading the manuscript. The research leading to these results has received funding from the European Community's Seventh Framework Program under grant agreement n. 259015 COLTHERES; AIRC 2010 Special Program Molecular Clinical Oncology 5xMille, Project n. 9970; Fondazione Piemontese per la Ricerca sul Cancro; Regione Piemonte.
Author information
Authors and Affiliations
Contributions
M. Martini, L. Vecchione and A. Bardelli researched the data for the article. All authors provided a substantial contribution to the discussion of the content. M. Martini, S. Tejpar and A. Bardelli wrote the manuscript and all authors edited and revised the article prior to submission.
Corresponding author
Ethics declarations
Competing interests
S. Tejpar declares that she receives research support from Merck Serono and Pfizer. The other authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Martini, M., Vecchione, L., Siena, S. et al. Targeted therapies: how personal should we go?. Nat Rev Clin Oncol 9, 87–97 (2012). https://doi.org/10.1038/nrclinonc.2011.164
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2011.164
This article is cited by
-
Patient Preferences in Targeted Pharmacotherapy for Cancers: A Systematic Review of Discrete Choice Experiments
PharmacoEconomics (2023)
-
AMPK activation overcomes anti-EGFR antibody resistance induced by KRAS mutation in colorectal cancer
Cell Communication and Signaling (2020)
-
A Novel System for Functional Determination of Variants of Uncertain Significance using Deep Convolutional Neural Networks
Scientific Reports (2020)
-
Restoring PUMA induction overcomes KRAS-mediated resistance to anti-EGFR antibodies in colorectal cancer
Oncogene (2018)
-
Reliance upon ancestral mutations is maintained in colorectal cancers that heterogeneously evolve during targeted therapies
Nature Communications (2018)