Key Points
-
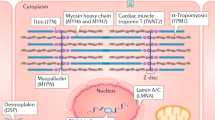
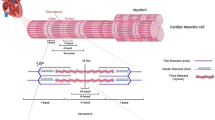
Titin-truncating variants (TTNtv) are strongly associated with dilated cardiomyopathy (DCM), but are also prevalent in the general population
-
TTNtv identified through genetic testing of patients with confirmed DCM might be clinically actionable and informative for the management of probands and their families
-
The relevance of TTNtv identified through sequencing for other indications is not well defined; such variants can be associated with increased risk of DCM, but in aggregate are not highly penetrant
-
Haploinsufficiency caused by TTNtv does not clearly explain all the associated molecular and physiological consequences, suggesting that other mechanisms also contribute to disease pathogenesis
-
Important genetic and environmental determinants of TTNtv penetrance and expressivity remain to be identified
Abstract
Dilated cardiomyopathy (DCM) affects approximately 1 in 250 individuals and is the leading indication for heart transplantation. DCM is often familial, and the most common genetic predisposition is a truncating variation in the giant sarcomeric protein, titin, which occurs in up to 15% of ambulant patients with DCM and 25% of end-stage or familial cases. In this article, we review the evidence for the role of titin truncation in the pathogenesis of DCM and our understanding of the molecular mechanisms and pathophysiological consequences of variation in the gene encoding titin (TTN). Such variation is common in the general population (up to 1% of individuals), and we consider key features that discriminate variants with disease-causing potential from those that are benign. We summarize strategies for clinical interpretation of genetic variants for use in the diagnosis of patients and the evaluation of their relatives. Finally, we consider the contemporary and potential future role for genetic stratification in cardiomyopathy and in the general population, evaluating titin variation as a predictor of outcome and treatment response for precision medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Linke, W. A. & Hamdani, N. Gigantic business. Circ. Res. 114, 1052–1068 (2014).
Tskhovrebova, L. & Trinick, J. Roles of titin in the structure and elasticity of the sarcomere. J. Biomed. Biotechnol. 2010, 1–7 (2010).
Linke, W. A. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 77, 637–648 (2008).
Gautel, M. The sarcomeric cytoskeleton: who picks up the strain? Curr. Opin. Cell Biol. 23, 39–46 (2011).
Puchner, E. M. et al. Mechanoenzymatics of titin kinase. Proc. Natl Acad. Sci. USA 105, 13385–13390 (2008).
Fürst, D. O., Osborn, M., Nave, R. & Weber, K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 106, 1563–1572 (1988).
Labeit, S. et al. A regular pattern of two types of 100-residue motif in the sequence of titin. Nature 345, 273–276 (1990).
Labeit, S. & Kolmerer, B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 270, 293–296 (1995).
Freiburg, A. et al. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res. 86, 1114–1121 (2000).
Luciano Brocchieri, S. K. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 33, 3390–3400 (2005).
Bang, M. L. et al. The complete gene sequence of titin, expression of an unusual 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 89, 1065–1072 (2001).
Neagoe, C., Opitz, C. A., Makarenko, I. & Linke, W. A. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J. Muscle Res. Cell Motil. 24, 175–189 (2003).
Neagoe, C. et al. Titin isoform switch in ischemic human heart disease. Circulation 106, 1333–1341 (2002).
Nagueh, S. F. et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation 110, 155–162 (2004).
Makarenko, I. et al. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 95, 708–716 (2004).
Borbély, A. et al. Hypophosphorylation of the stiff N2B titin isoform raises cardiomyocyte resting tension in failing human myocardium. Circ. Res. 104, 780–786 (2009).
Opitz, C. A. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ. Res. 94, 967–975 (2004).
Zou, J. et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of titin in zebrafish. eLife 4, 1065–1036 (2015).
Schafer, S. et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 49, 46–53 (2017).
Pinto, Y. M. et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur. Heart J. 14, 1850–1858 (2016).
Hershberger, R. E., Hedges, D. J. & Morales, A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 10, 531–547 (2013).
Elliott, P. et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 29, 270–276 (2008).
Herman, D. S. et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366, 619–628 (2012).
Ware, J. S. et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N. Engl. J. Med. 374, 233–241 (2016).
Køber et al. Defibrillator implantation in patients with nonischemic systolic heart failure. N. Engl. J. Med. 375, 1221–1230 (2016).
Gulati, A. et al. Association of fibrosis with mortality and sudden cardiac death in patients with nonischemic dilated cardiomyopathy. JAMA 309, 896–908 (2013).
Maron, B. J. et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 113, 1807–1816 (2006).
Stehlik, J. et al. The registry of the International Society for Heart and Lung Transplantation: Twenty-eighth Adult Heart Transplant Report — 2011. J. Heart Lung Transplant. 30, 1078–1094 (2011).
Haravuori, H. et al. Assignment of the tibial muscular dystrophy locus to chromosome 2q31. Am. J. Hum. Genet. 62, 620–626 (2017).
Siu, B. L. et al. Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation 99, 1022–1026 (1999).
Gerull, B. et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 30, 201–204 (2002).
Xu, X. et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 30, 205–209 (2002).
Gerull, B. et al. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J. Mol. Med. 84, 478–483 (2006).
Carmignac, V. et al. C-Terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann. Neurol. 61, 340–351 (2007).
Norton, N. et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ. Cardiovasc. Genet. 6, 144–153 (2013).
Pugh, T. J. et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet. Med. 16, 601–608 (2014).
van Spaendonck-Zwarts, K. Y. et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur. Heart J. 35, 2165–2173 (2014).
Roberts, A. M. et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl Med. 7, 270ra6 (2015).
Haas, J. et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 36, 1123–1135 (2015).
Akinrinade, O. et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 36, 2327–2337 (2015).
Walsh, R. et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 19, 192–203 (2017).
Franaszczyk, M. et al. Titin truncating variants in dilated cardiomyopathy — prevalence and genotype-phenotype correlations. PLoS ONE 12, e0169007 (2017).
Tayal, U. et al. Phenotype and clinical outcomes of titin cardiomyopathy. J. Am. Coll. Cardiol. 18, 2264–2274 (2017).
Fatkin, D. et al. Titin truncating mutations: a rare cause of dilated cardiomyopathy in the young. Prog. Pediatr. Cardiol. 40, 41–45 (2016).
van Spaendonck-Zwarts, K. Y. et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation 121, 2169–2175 (2010).
Hastings, R. et al. Combination of whole genome sequencing, linkage, and functional studies implicates a missense mutation in titin as a cause of autosomal dominant cardiomyopathy with features of left ventricular noncompaction. Circ. Cardiovasc. Genet. 9, 426–435 (2016).
Ceyhan-Birsoy, O. et al. Recessive truncating titin gene, TTN, mutations presenting as centronuclear myopathy. Neurology 81, 1205–1214 (2013).
Chauveau, C. et al. Recessive TTN truncating mutations define novel forms of core myopathy with heart disease. Hum. Mol. Genet. 23, 980–991 (2014).
Pfeffer, G. et al. Titin founder mutation is a common cause of myofibrillar myopathy with early respiratory failure. J. Neurol. Neurosurg. Psychiatry 85, 331–338 (2014).
Palmio, J. et al. Hereditary myopathy with early respiratory failure: occurrence in various populations. J. Neurol. Neurosurg. Psychiatry 85, 345–353 (2014).
Fernández-Marmiesse, A. et al. Homozygous truncating mutation in prenatally expressed skeletal isoform of TTN gene results in arthrogryposis multiplex congenita and myopathy without cardiac involvement. Neuromuscul. Disord. 27, 188–192 (2017).
Chauveau, C., Rowell, J. & Ferreiro, A. A rising titan: TTN review and mutation update. Hum. Mutat. 35, 1046–1059 (2014).
Lopes, L. R. et al. Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J. Med. Genet. 50, 228–239 (2013).
Satoh, M. et al. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem. Biophys. Res. Commun. 262, 411–417 (1999).
Arimura, T. et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 54, 334–342 (2009).
Li, S., Guo, W., Dewey, C. N. & Greaser, M. L. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 41, 2659–2672 (2013).
Guo, W. et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18, 766–773 (2012).
Beqqali, A. et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc. Res. 112, 452–463 (2016).
LeWinter, M. M. & Granzier, H. L. Titin is a major human disease gene. Circulation 127, 938–944 (2013).
Watkins, H. et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 11, 434–437 (1995).
Bonne, G. et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat. Genet. 11, 438–440 (1995).
Redwood, C. Properties of mutant contractile proteins that cause hypertrophic cardiomyopathy. Cardiovasc. Res. 44, 20–36 (1999).
Marston, S. et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ. Res. 105, 219–222 (2009).
Hinson, J. T. et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986 (2015).
Harding, S. E., MacLeod, K. T., Jones, S. M., Vescovo, G. & Poole-Wilson, P. A. Contractile responses of myocytes isolated from patients with cardiomyopathy. Eur. Heart J. 12 (Suppl. D), 44–48 (1991).
Song, W. et al. Investigation of a transgenic mouse model of familial dilated cardiomyopathy. J. Mol. Cell. Cardiol. 49, 380–389 (2010).
Stanley, W. C., Recchia, F. A. & Lopaschuk, G. D. Myocardial substrate metabolism in the normal and failing heart. Physiol. Rev. 85, 1093–1129 (2005).
Doenst, T., Nguyen, T. D. & Abel, E. D. Cardiac metabolism in heart failure: implications beyond ATP production. Circ. Res. 113, 709–724 (2013).
Lai, L. et al. Energy metabolic reprogramming in the hypertrophied and early stage failing heart: a multisystems approach. Circ. Heart Fail. 7, 1022–1031 (2014).
Shibayama, J. et al. Metabolic remodeling in moderate synchronous versus dyssynchronous pacing-induced heart failure: integrated metabolomics and proteomics study. PLoS ONE 10, e0118974 (2015).
Schisler, J. C. et al. Cardiac energy dependence on glucose increases metabolites related to glutathione and activates metabolic genes controlled by mechanistic target of rapamycin. J. Am. Heart Assoc. 4, e001136 (2015).
Neishabouri, S. H., Hutson, S. M. & Davoodi, J. Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino Acids 47, 1167–1182 (2015).
Ramos, F. J. et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl Med. 4, 144ra103 (2012).
Yano, T. et al. Clinical impact of myocardial mTORC1 activation in nonischemic dilated cardiomyopathy. J. Mol. Cell. Cardiol. 91, 6–9 (2016).
Radke, M. H. et al. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc. Natl Acad. Sci. USA 104, 3444–3449 (2007).
Lunde, I. G. et al. A deletion in the N2A region of titin carried by muscular dystrophy with myositis (mdm) mice severely affects skeletal muscle, but not the heart [abstract]. Circ. Res. 115 (Suppl. 1), A279 (2014).
Gramlich, M. et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J. Mol. Cell. Cardiol. 47, 352–358 (2009).
Lunde, I. G. et al. Titin A-band truncation in mice causes stress-induced dilated cardiomyopathy [abstract]. Presented at the 14th Annual Center for Heart Failure Research Symposium on Heart Failure (2016).
Jansweijer, J. A. et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur. J. Heart Fail. 19, 512–521 (2016).
Tayal, U. et al. Truncating variants in titin independently predict early arrhythmias in patients with dilated cardiomyopathy. J. Am. Coll. Cardiol. 69, 2466–2468 (2017).
Ackerman, M. J. et al. HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for the Channelopathies and Cardiomyopathies: This document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 13, 1077–1109 (2011).
Charron, P. et al. Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 31, 2715–2726 (2010).
McMurray, J. J. V. et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 33, 1787–1847 (2012).
Yancy, C. W. et al. 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 128, e240–e327 (2013).
Akinrinade, O., Koskenvuo, J. W. & Alastalo, T.-P. Prevalence of titin truncating variants in general population. PLoS ONE 10, e0145284 (2015).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423 (2015).
Minikel, E. V. et al. Quantifying prion disease penetrance using large population control cohorts. Sci. Transl Med. 8, 322ra9 (2016).
Whiffin, N. et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. 19, 1151–1158 (2017).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Rivas, M. A. et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348, 666–669 (2015).
Holbrook, J. A., Neu-Yilik, G., Hentze, M. W. & Kulozik, A. E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36, 801–808 (2004).
Nagy, E. & Maquat, L. E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998).
Deo, R. C. Alternative splicing, internal promoter, nonsense-mediated decay, or all three. Circ. Cardiovasc. Genet. 9, 419–425 (2016).
Van Buggenhout, G. et al. The del(2)(q32.2q33) deletion syndrome defined by clinical and molecular characterization of four patients. Eur. J. Med. Genet. 48, 276–289 (2005).
Prontera, P., Bernardini, L. & Dallapiccola, B. 2q31.2q32.3 deletion syndrome: report of an adult patient. Am. J. Med. Genet. A 149A, 706–712 (2009).
Mencarelli, M. A. et al. Clinical and molecular characterization of a patient with a 2q31.2-32.3 deletion identified by array-CGH. Am. J. Med. Genet. A 143A, 858–865 (2007).
Rifai, L. et al. Ectodermal dysplasia-like syndrome with mental retardation due to contiguous gene deletion: Further clinical and molecular delineation of del(2q32) syndrome. Am. J. Med. Genet. A 152A, 111–117 (2009).
Mitter, D. et al. Genotype-phenotype correlation in eight new patients with a deletion encompassing 2q31.1. Am. J. Med. Genet. A 152A, 1213–1224 (2010).
Manolakos, E. et al. Deletion 2q31.2-q31.3 in a 4-year-old girl with microcephaly and severe mental retardation. Am. J. Med. Genet. 155, 1476–1482 (2011).
Taylor, M. R. G. et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J. Am. Coll. Cardiol. 41, 771–780 (2003).
Berlo, J. H. et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. 83, 79–83 (2004).
Felkin, L. E. et al. Recovery of cardiac function in cardiomyopathy caused by titin truncation. JAMA Cardiol. 1, 234–235 (2016).
Luk, K. et al. Recovery in patients with dilated cardiomyopathy with loss-of-function mutations in the titin gene. JAMA Cardiol. 2, 700–702 (2017).
Mann, D. L., Barger, P. M. & Burkhoff, D. Myocardial recovery and the failing heart: myth, magic, or molecular target? J. Am. Coll. Cardiol. 60, 2465–2472 (2012).
Linschoten, M. et al. Truncating titin (TTN) variants in chemotherapy-induced cardiomyopathy. J. Card. Fail. 23, 476–479 (2017).
Gramlich, M. et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 7, 562–576 (2015).
Aartsma-Rus, A. & Krieg, A. M. FDA approves eteplirsen for Duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid. Ther. 27, 1–3 (2017).
Wilkie, A. O. The molecular basis of genetic dominance. J. Med. Genet. 31, 89–98 (1994).
Acknowledgements
The authors thank Zofia T. Bilinska (Cardinal Stefan Wyszynski Institute of Cardiology, Warsaw, Poland) for providing access to the data for Figure 4, and acknowledge support from the British Heart Foundation; Fondation Leducq; Medical Research Council, UK; National Medical Research Council Singapore; National Institute for Health Research (NIHR) Imperial Biomedical Research Centre; NIHR Royal Brompton Biomedical Research Unit; SingHealth Duke–National University Singapore (Duke–NUS) Institute of Precision Medicine; and Wellcome Trust (107469/Z/15/Z).
Author information
Authors and Affiliations
Contributions
J.S.W. and S.A.C. researched data for the article, discussed the content, wrote the manuscript, and reviewed and/or edited the article before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Assigning risk and causality to genetic variants. (PDF 122 kb)
Rights and permissions
About this article
Cite this article
Ware, J., Cook, S. Role of titin in cardiomyopathy: from DNA variants to patient stratification. Nat Rev Cardiol 15, 241–252 (2018). https://doi.org/10.1038/nrcardio.2017.190
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrcardio.2017.190
This article is cited by
-
Genetic characterization of dilated cardiomyopathy patients undergoing heart transplantation in the Chinese population by whole-exome sequencing
Journal of Translational Medicine (2023)
-
Environmental and genetic predictors of human cardiovascular ageing
Nature Communications (2023)
-
Tools to differentiate between Filamin C and Titin truncating variant carriers: value of MRI
European Journal of Human Genetics (2023)
-
Targeting the sarcomere in inherited cardiomyopathies
Nature Reviews Cardiology (2022)
-
Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments
Heart Failure Reviews (2022)