Key Points
-
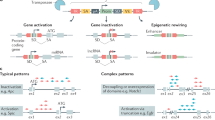
Transposon-based insertional mutagenesis (TIM) can be used to model many types of cancer in mice and facilitates the rapid identification of genes that cause cancer.
-
Comparison of the genes mutated by TIM with the genes mutated in human cancer can help to discriminate between those that are driver and those that are passenger mutations in human cancer.
-
TIM can identify new human cancer genes that have so far been missed in human cancer genome sequencing studies, that are difficult to identify because they are epigenetically regulated and that are located in large amplified or deleted regions.
-
TIM induces tumours at all stages of tumour progression, sometimes in a single animal. By identifying the genes mutated by TIM at each stage of progression it might be possible to determine the order in which the mutations were acquired and whether they are involved in tumour initiation, progression and metastasis.
-
Mouse cancer models induced by TIM provide a plethora of new models for studying the biology of cancer and for testing new cancer therapeutics before they go into the clinic.
Abstract
Recently, it has become possible to mobilize the Tc1/mariner transposon, Sleeping Beauty (SB), in mouse somatic cells at frequencies high enough to induce cancer. Tumours result from SB insertional mutagenesis of cancer genes, thus facilitating the identification of the genes and signalling pathways that drive tumour formation. A conditional SB transposition system has also been developed that makes it possible to limit where SB mutagenesis occurs, providing a means to selectively model many types of human cancer. SB mutagenesis has already identified a large collection of known cancer genes in addition to a plethora of new candidate cancer genes and potential drug targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Imai, K. & Takaoka, A. Comparing antibody and small-molecule therapies for cancer. Nature Rev. Cancer 6, 714–727 (2006).
Copeland, N. G. & Jenkins, N. A. Deciphering the genetic landscape of cancer-from genes to pathways. Trends Genet. 25, 455–462 (2009).
Forrest, W. F. & Cavet, G. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
Getz, G. et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
Rubin, A. F. & Green, P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007).
Kool, J. & Berns, A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nature Rev. Cancer 9, 389–399 (2009). This paper reviews the advantages and the limitations of retroviral insertional mutagenesis in mice for identifying genes and signalling pathways that are important for cancer.
Akagi, K., Suzuki, T., Stephens, R. M., Jenkins, N. A. & Copeland, N. G. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 32, D523–D527 (2004).
Mitchell, R. S. et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2, e234 (2004).
Wu, X., Li, Y., Crise, B. & Burgess, S. M. Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 (2003).
Neil, J. C. & Cameron, E. R. Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2, 253–255 (2002).
Plasterk, R. H., Izsvak, Z. & Ivics, Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15, 326–332 (1999).
Lohe, A. R., Moriyama, E. N., Lidholm, D. A. & Hartl, D. L. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12, 62–72 (1995).
Izsvak, Z., Ivics, Z. & Hackett, P. B. Repetitive elements and their genetic applications in zebrafish. Biochem. Cell Biol. 75, 507–523 (1997).
Ivics, Z., Izsvak, Z., Minter, A. & Hackett, P. B. Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl Acad. Sci. USA 93, 5008–5013 (1996).
Zayed, H., Izsvak, Z., Walisko, O. & Ivics, Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol. Ther. 9, 292–304 (2004).
Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 (1997). A landmark paper that constructed the first synthetic highly active transposon (SB) for use in mammalian cells.
Izsvak, Z., Ivics, Z. & Plasterk, R. H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302, 93–102 (2000).
Yant, S. R. et al. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nature Genet. 25, 35–41 (2000).
Luo, G., Ivics, Z., Izsvak, Z. & Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 95, 10769–10773 (1998).
Dupuy, A. J. et al. Mammalian germ-line transgenesis by transposition. Proc. Natl Acad. Sci. USA 99, 4495–4499 (2002).
Dupuy, A. J., Fritz, S. & Largaespada, D. A. Transposition and gene disruption in the male germline of the mouse. Genesis 30, 82–88 (2001).
Fischer, S. E., Wienholds, E. & Plasterk, R. H. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl Acad. Sci. USA 98, 6759–6764 (2001).
Horie, K. et al. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc. Natl Acad. Sci. USA 98, 9191–9196 (2001).
Zayed, H., Izsvak, Z., Khare, D., Heinemann, U. & Ivics, Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31, 2313–2322 (2003).
Walisko, O. et al. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl Acad. Sci. USA 103, 4062–4067 (2006).
Izsvak, Z. et al. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13, 279–290 (2004).
Yant, S. R. & Kay, M. A. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol. Cell. Biol. 23, 8505–8518 (2003).
Ikeda, R. et al. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol. Cell. Biol. 27, 1665–1676 (2007).
Yusa, K., Takeda, J. & Horie, K. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol. Cell. Biol. 24, 4004–4018 (2004).
Garrison, B. S., Yant, S. R., Mikkelsen, J. G. & Kay, M. A. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol. Cell. Biol. 27, 8824–8833 (2007).
Park, C. W., Kren, B. T., Largaespada, D. A. & Steer, C. J. DNA methylation of Sleeping Beauty with transposition into the mouse genome. Genes Cells 10, 763–776 (2005).
Park, C. W., Park, J., Kren, B. T. & Steer, C. J. Sleeping Beauty transposition in the mouse genome is associated with changes in DNA methylation at the site of insertion. Genomics 88, 204–213 (2006).
Walsh, C. P. & Bestor, T. H. Cytosine methylation and mammalian development. Genes Dev. 13, 26–34 (1999).
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Liu, G. et al. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 346, 161–173 (2005).
Vigdal, T. J., Kaufman, C. D., Izsvak, Z., Voytas, D. F. & Ivics, Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323, 441–452 (2002).
Yant, S. R. et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 25, 2085–2094 (2005).
Carlson, C. M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Horie, K. et al. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23, 9189–9207 (2003).
Tower, J., Karpen, G. H., Craig, N. & Spradling, A. C. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133, 347–359 (1993).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226 (2005). This was the first paper showing that SB transposon-based mutagenesis can induce cancer when mobilized in somatic cells of wild-type mice.
Geurts, A. M. et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol. Ther. 8, 108–117 (2003).
Cui, Z., Geurts, A. M., Liu, G., Kaufman, C. D. & Hackett, P. B. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 318, 1221–1235 (2002).
Liang, Q., Kong, J., Stalker, J. & Bradley, A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47, 404–408 (2009).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999).
Zambrowicz, B. P. et al. Disruption of overlapping transcripts in the ROSA β geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA 94, 3789–3794 (1997).
Geurts, A. M. et al. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2, e156 (2006).
Ellisen, L. W. et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991).
Collier, L. S., Carlson, C. M., Ravimohan, S., Dupuy, A. J. & Largaespada, D. A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436, 272–276 (2005). This was the first paper showing that SB transposon-based mutagenesis can identify genes involved in solid cancer when mobilized in somatic cells of sensitized mice.
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).
Seidel, C. et al. Alterations of cancer-related genes in soft tissue sarcomas: hypermethylation of RASSF1A is frequently detected in leiomyosarcoma and associated with poor prognosis in sarcoma. Int. J. Cancer 114, 442–447 (2005).
Collier, L. S. et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 69, 8429–8437 (2009).
Rahrmann, E. P. et al. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 69, 4388–4397 (2009).
Hawley, R. G., Fong, A. Z., Burns, B. F. & Hawley, T. S. Transplantable myeloproliferative disease induced in mice by an interleukin 6 retrovirus. J. Exp. Med. 176, 1149–1163 (1992).
Dupuy, A. J. et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 69, 8150–8156 (2009). This paper describes the development of a modified SB transposon, T2/Onc3, which selectively induces epithelial tumours when mobilized in somatic cells of wild-type mice.
Niwa, H., Yamamura, K. & Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991).
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
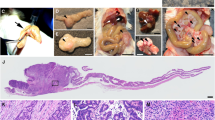
Keng, V. W. et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nature Biotech. 27, 264–274 (2009). This paper describes the development of a conditional transposition system that can be used to model many different kinds of human cancer in mice. This transposition system was also used in a proof-of-principle experiment to model HCC in mice.
Starr, T. K. et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323, 1747–1750 (2009). This paper describes the development of a conditional transposition system that can be used to model many different kinds of human cancer in mice. This transposition system was also used in a proof-of-principle experiment to model CRC in mice.
Segditsas, S. & Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 25, 7531–7537 (2006).
Rajagopalan, H. et al. Inactivation of hCDC4 can cause chromosomal instability. Nature 428, 77–81 (2004).
de Vries, A. et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc. Natl Acad. Sci. USA 99, 2948–2953 (2002).
Bressac, B., Kew, M., Wands, J. & Ozturk, M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350, 429–431 (1991).
Buendia, M. A. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 10, 185–200 (2000).
Boerner, J. L., Danielsen, A. & Maihle, N. J. Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp. Cell Res. 284, 111–121 (2003).
Chang, C. M. et al. A minor tyrosine phosphorylation site located within the CAIN domain plays a critical role in regulating tissue-specific transformation by erbB kinase. J. Virol. 69, 1172–1180 (1995).
Frederick, L., Wang, X. Y., Eley, G. & James, C. D. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 60, 1383–1387 (2000).
Villanueva, A. et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 135, 1972–1983 (2008).
Villanueva, A., Newell, P., Chiang, D. Y., Friedman, S. L. & Llovet, J. M. Genomics and signaling pathways in hepatocellular carcinoma. Semin. Liver Dis. 27, 55–76 (2007).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Salvi, A. et al. In vitro c-met inhibition by antisense RNA and plasmid-based RNAi down-modulates migration and invasion of hepatocellular carcinoma cells. Int. J. Oncol. 31, 451–460 (2007).
Takami, T. et al. Loss of hepatocyte growth factor/c-Met signaling pathway accelerates early stages of N-nitrosodiethylamine induced hepatocarcinogenesis. Cancer Res. 67, 9844–9851 (2007).
Nakayama, K. et al. Homozygous deletion of MKK4 in ovarian serous carcinoma. Cancer Biol. Ther. 5, 630–634 (2006).
Su, G. H., Song, J. J., Repasky, E. A., Schutte, M. & Kern, S. E. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum. Mutat. 19, 81 (2002).
Xin, W. et al. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin. Cancer Res. 10, 8516–8520 (2004).
Arsura, M. et al. Transient activation of NF-κB through a TAK1/IKK kinase pathway by TGF-β1 inhibits AP-1/SMAD signaling and apoptosis: implications in liver tumor formation. Oncogene 22, 412–425 (2003).
Anzola, M. Hepatocellular carcinoma: role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J. Viral Hepat. 11, 383–393 (2004).
Beasley, R. P. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61, 1942–1956 (1988).
Perz, J. F., Armstrong, G. L., Farrington, L. A., Hutin, Y. J. & Bell, B. P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45, 529–538 (2006).
Yang, H. I. et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N. Engl. J. Med. 347, 168–174 (2002).
Mizuuchi, K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell 35, 785–794 (1983).
de Wit, T. et al. Tagged mutagenesis by efficient Minos-based germ line transposition. Mol. Cell. Biol. 30, 68–77 (2010).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005). This paper shows that PB is a highly active transposon in mammalian cells and the mouse germ line and an excellent alternative transposition system to SB for insertional mutagenesis screens in mice.
Keng, V. W. et al. Efficient transposition of Tol2 in the mouse germline. Genetics 183, 1565–1573 (2009).
Wang, W. et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 9290–9295 (2008).
Smith, G. E., Summers, M. D. & Fraser, M. J. Production of human β interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 3, 2156–2165 (1983).
Fraser, M. J., Cary, L., Boonvisudhi, K. & Wang, H. G. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology 211, 397–407 (1995).
Fraser, M. J., Ciszczon, T., Elick, T. & Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5, 141–151 (1996).
Cadinanos, J. & Bradley, A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 35, e87 (2007).
Mates, L. et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nature Genet. 41, 753–761 (2009).
Grabundzija, I. et al. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 18, 1200–1209 (2010).
Chambraud, B., Berry, M., Redeuilh, G., Chambon, P. & Baulieu, E. E. Several regions of human estrogen receptor are involved in the formation of receptor-heat shock protein 90 complexes. J. Biol. Chem. 265, 20686–20691 (1990).
Feil, R., Wagner, J., Metzger, D. & Chambon, P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237, 752–757 (1997).
Mitra, R., Fain-Thornton, J. & Craig, N. L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 27, 1097–1109 (2008).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 17, 688–695 (2010).
Lauchle, J. O. et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature 461, 411–414 (2009).
de Ridder, J., Uren, A., Kool, J., Reinders, M. & Wessels, L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput. Biol. 2, e166 (2006).
Uren, A. G. et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell 133, 727–741 (2008). This paper shows how large-scale screens in mice carrying sensitizing cancer causing mutations can be used to identify genes that are involved in collaborative cancer networks.
Yang, Y. H. et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15 (2002).
Devon, R. S., Porteous, D. J. & Brookes, A. J. Splinkerettes-improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23, 1644–1645 (1995).
Gavin, A. J. et al. Pooled library tissue tags for EST-based gene discovery. Bioinformatics 18, 1162–1166 (2002).
Acknowledgements
The authors would like to thank S. C. Lee for help with the figures and members of their laboratory for helpful comments on the Review. Support was provided by the Biomedical Research Council (BMRC), Agency for Science and Technology and Research (A*STAR), Singapore.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
FURTHER INFORMATION
Glossary
- Preputial
-
Exocrine glands that produce pheromones found in front of the genitals in some mammals.
- Ingenuity pathway analysis
-
A curated database and analysis system used to explain how proteins work together to cause cellular changes.
- Kernel function
-
A weighting function used in nonparametric function estimation. It gives the weights of the nearby data points in making an estimate.
- Bonferroni correction
-
A method used to address the problem of multiple comparisons.
Rights and permissions
About this article
Cite this article
Copeland, N., Jenkins, N. Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10, 696–706 (2010). https://doi.org/10.1038/nrc2916
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2916
This article is cited by
-
Sleeping Beauty transposon mutagenesis in mouse intestinal organoids identifies genes involved in tumor progression and metastasis
Cancer Gene Therapy (2024)
-
Sleeping Beauty transposon mutagenesis identified genes and pathways involved in inflammation-associated colon tumor development
Nature Communications (2023)
-
Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis
Communications Biology (2020)
-
Haematopoiesis in the era of advanced single-cell technologies
Nature Cell Biology (2019)
-
Insertional mutagenesis using the Sleeping Beauty transposon system identifies drivers of erythroleukemia in mice
Scientific Reports (2019)