Abstract
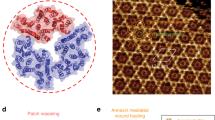
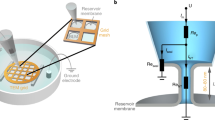
Annexins are abundant cytoplasmic proteins that can bind to negatively charged phospholipids in a Ca2+-dependent manner, and are known to play a role in the storage of Ca2+ and membrane healing. Little is known, however, about the dynamic processes of protein–Ca2+–membrane assembly and disassembly. Here we show that high-speed atomic force microscopy (HS-AFM) can be used to repeatedly induce and disrupt annexin assemblies and study their structure, dynamics and interactions. Our HS-AFM set-up is adapted for such biological applications through the integration of a pumping system for buffer exchange and a pulsed laser system for uncaging caged compounds. We find that biochemically identical annexins (annexin V) display different effective Ca2+ and membrane affinities depending on the assembly location, providing a wide Ca2+ buffering regime while maintaining membrane stabilization. We also show that annexin is membrane-recruited and forms stable supramolecular assemblies within ∼5 s in conditions that are comparable to a membrane lesion in a cell. Molecular dynamics simulations provide atomic detail of the role played by Ca2+ in the reversible binding of annexin to the membrane surface.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gerke, V. & Moss, S. E. Annexins: from structure to function. Physiol. Rev. 82, 331–371 (2002).
Gauer Jacob, W. et al. Membrane modulates affinity for calcium ion to create an apparent cooperative binding response by annexin a5. Biophys. J. 104, 2437–2447 (2013).
Lizarbe, M., Barrasa, J., Olmo, N., Gavilanes, F. & Turnay, J. Annexin-phospholipid interactions. Functional implications. Int. J. Mol. Sci. 14, 2652–2683 (2013).
Gerke, V., Creutz, C. E. & Moss, S. E. Annexins: linking Ca2+ signalling to membrane dynamics. Nature Rev. Mol. Cell Biol. 6, 449–461 (2005).
Bouter, A. et al. Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nature Commun. 2, 274–279 (2011).
McNeil, A. K., Rescher, U., Gerke, V. & McNeil, P. L. Requirement for annexin A1 in plasma membrane repair. J. Biol. Chem. 281, 35202–35207 (2006).
Vermes, I., Haanen, C., Steffens-Nakken, H. & Reutellingsperger, C. A novel assay for apoptosis flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J. Immun. Meth. 184, 39–51 (1995).
Bewley, M. C., Boustead, C. M., Walker, J. H., Waller, D. A. & Huber, R. Structure of chicken annexin-V at 2.25-A resolution. Biochemistry 32, 3923–3929 (1993).
Meers, P. in Annexins: Molecular Structure to Cellular Function (ed. Seaton, B. A.) 97–119 (RG Landes Company, 1996).
Concha, N. O., Head, J. F., Kaetzel, M. A., Dedman, J. R. & Seaton, B. A. Annexin V forms calcium-dependent trimeric units on phospholipid vesicles. FEBS Lett. 314, 159–162 (1992).
Voges, D. et al. Three-dimensional structure of membrane-bound annexin V. A correlative electron microscopy-X-ray crystallography study. J. Mol. Biol. 238, 199–213 (1994).
Mosser, G., Ravanat, C., Freyssinet, J. M. & Brisson, A. Sub-domain structure of lipid-bound annexin-V resolved by electron image analysis. J. Mol. Biol. 217, 241–245 (1991).
Brisson, A., Olofsson, A., Ringler, P., Schmutz, M. & Stoyla, S. Two-dimensional crystallization of proteins on planar lipid films and structure determination by electron crystallography. Biol. Cell. 80, 221–228 (1994).
Oling, F., Bergsma-Schutter, W. & Brisson, A. Trimers, dimers of trimers, and trimers of trimers are common building blocks of annexin A5 two-dimensional crystals. J. Struct. Biol. 133, 55–63 (2001).
Reviakine, I., Bergsma-Schutter, W. & Brisson, A. Growth of protein 2-D crystals on supported planar lipid bilayers imaged in situ by AFM. J. Struct. Biol. 121, 356–361 (1998).
Pigault, C., Follenius-Wund, A., Schmutz, M., Freyssinet, J.-M. & Brisson, A. Formation of two-dimensional arrays of annexin V on phosphatidylserine-containing liposomes. J. Mol. Biol. 236, 199–208 (1994).
Langen, R., Isas, J. M., Luecke, H., Haigler, H. T. & Hubbell, W. L. Membrane-mediated assembly of annexins studied by site-directed spin labeling. J. Biol. Chem. 273, 22453–22457 (1998).
Ando, T. et al. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl Acad. Sci. USA 98, 12468–12472 (2001).
Ando, T., Uchihashi, T. & Scheuring, S. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 114, 3120–3188 (2014).
Patel, D. R. et al. The conserved core domains of annexins A1, A2, A5, and B12 can be divided into two groups with different Ca2+-dependent membrane-binding properties. Biochemistry 44, 2833–2844 (2005).
Casuso, I. et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nature Nanotech. 7, 525–529 (2012).
Kaplan, J. H. & Ellis-Davies, G. C. Photolabile chelators for the rapid photorelease of divalent cations. Proc. Natl Acad. Sci. USA 85, 6571–6575 (1988).
Husain, M., Boudier, T., Paul-Gilloteaux, P., Casuso, I. & Scheuring, S. Software for drift compensation, particle tracking and particle analysis of high-speed atomic force microscopy image series. J. Mol. Rec. 25, 292–298 (2012).
Yamamoto, D., Uchihashi, T., Kodera, N. & Ando, T. Anisotropic diffusion of point defects in a two-dimensional crystal of streptavidin observed by high-speed atomic force microscopy. Nanotechnology 19, 384009 (2008).
Fechner, P. et al. Structural information, resolution, and noise in high-resolution atomic force microscopy topographs. Biophys. J. 96, 3822–3831 (2009).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comp. Chem. 26, 1781–1802 (2005).
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102, 3586–3616 (1998).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 114, 7830–7843 (2010).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Miyamoto, S. & Kollman, P. A. Settle: an analytical version of the shake and rattle algorithm for rigid water models. J. Comp. Chem. 13, 952–962 (1992).
Acknowledgements
The authors thank H. Haigler for sharing insights in annexin biophysics and structural biology and for valuable comments on the manuscript, and A. Karner for assistance with the setting up of the fluid exchange system. This work was funded by the ANR grants ANR-Nano (ANR-12-BS10-009-01) and ANR-BBMS (ANR-12-BSV8-0006-01) and a European Research Council (ERC) Grant (No. 310080). The GENCI and CINES, Montpellier, France, are acknowledged for the provision of computer time.
Author information
Authors and Affiliations
Contributions
S.S., A.M and C.C conceived and designed the experiments. A.M., C.C. and M.R. performed the experiments. S.S., C.C and A.M analysed the data. S.S., C.C and A.M wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 435 kb)
Supplementary information
Supplementary Movie 1 (GIF 4602 kb)
Supplementary information
Supplementary Movie 2 (GIF 6342 kb)
Supplementary information
Supplementary Movie 3 (GIF 1283 kb)
Supplementary information
Supplementary Movie 4 (GIF 50128 kb)
Supplementary information
Supplementary Movie 5 (GIF 50957 kb)
Supplementary information
Supplementary Movie 6 (GIF 27022 kb)
Supplementary information
Supplementary Movie 7 (GIF 56262 kb)
Rights and permissions
About this article
Cite this article
Miyagi, A., Chipot, C., Rangl, M. et al. High-speed atomic force microscopy shows that annexin V stabilizes membranes on the second timescale. Nature Nanotech 11, 783–790 (2016). https://doi.org/10.1038/nnano.2016.89
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2016.89
This article is cited by
-
Molecular characterization and functional implications on mouse peripheral blood mononuclear cells of annexin proteins from Echinococcus granulosus sensu lato
Parasites & Vectors (2023)
-
Technical advances in high-speed atomic force microscopy
Biophysical Reviews (2023)
-
Perforin-2 clockwise hand-over-hand pre-pore to pore transition mechanism
Nature Communications (2022)
-
Interplay of membrane crosslinking and curvature induction by annexins
Scientific Reports (2022)