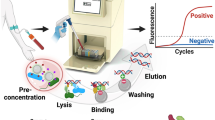
Abstract
So far, although various diagnostic approaches for pathogen detection have been proposed, most are too expensive, lengthy or limited in specificity for clinical use. Nanoparticle systems with unique material properties, however, circumvent these problems and offer improved accuracy over current methods. Here, we present novel magneto-DNA probes capable of rapid and specific profiling of pathogens directly in clinical samples. A nanoparticle hybridization assay, involving ubiquitous and specific probes that target bacterial 16S rRNAs, was designed to detect amplified target DNAs using a miniaturized NMR device. Ultimately, the magneto-DNA platform will allow both universal and specific detection of various clinically relevant bacterial species, with sensitivity down to single bacteria. Furthermore, the assay is robust and rapid, simultaneously diagnosing a panel of 13 bacterial species in clinical specimens within 2 h. The generic platform described could be used to rapidly identify and phenotype pathogens for a variety of applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Allegranzi, B. et al. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet 377, 228–241 (2011).
Polin, R. A. et al. Epidemiology and diagnosis of health care-associated infections in the NICU. Pediatrics 129, e1104–e1109 (2012).
Klompas, M., Yokoe, D. S. & Weinstein, R. A. Automated surveillance of health care-associated infections. Clin. Infect. Dis. 48, 1268–1275 (2009).
Giljohann, D. A. & Mirkin, C. A. Drivers of biodiagnostic development. Nature 462, 461–464 (2009).
Loman, N. J. et al. Performance comparison of benchtop high-throughput sequencing platforms. Nature Biotechnol. 30, 434–439 (2012).
Sauer, S. & Kliem, M. Mass spectrometry tools for the classification and identification of bacteria. Nature Rev. Microbiol. 8, 74–82 (2010).
Li, Y., Cu, Y. T. & Luo, D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nature Biotechnol. 23, 885–889 (2005).
Dark, P. M., Dean, P. & Warhurst, G. Bench-to-bedside review: the promise of rapid infection diagnosis during sepsis using polymerase chain reaction-based pathogen detection. Crit. Care 13, 217 (2009).
Pechorsky, A., Nitzan, Y. & Lazarovitch, T. Identification of pathogenic bacteria in blood cultures: comparison between conventional and PCR methods. J. Microbiol. Methods 78, 325–330 (2009).
Ottesen, E. A., Hong, J. W., Quake, S. R. & Leadbetter, J. R. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467 (2006).
Loman, N. J. et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nature Rev. Microbiol. 10, 599–606 (2012).
Niemz, A., Ferguson, T. M. & Boyle, D. S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29, 240–250 (2011).
Ward, D. M., Weller, R. & Bateson, M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345, 63–65 (1990).
Yang, S. et al. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J. Clin. Microbiol. 40, 3449–3454 (2002).
Rajendhran, J. & Gunasekaran, P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol. Res. 166, 99–110 (2011).
Woo, P. C., Lau, S. K., Teng, J. L., Tse, H. & Yuen, K. Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14, 908–934 (2008).
Lee, H., Sun, E., Ham, D. & Weissleder, R. Chip-NMR biosensor for detection and molecular analysis of cells. Nature Med. 14, 869–874 (2008).
Haun, J. B., Devaraj, N. K., Hilderbrand, S. A., Lee, H. & Weissleder, R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nature Nanotech. 5, 660–665 (2010).
Lee, H., Yoon, T. J., Figueiredo, J. L., Swirski, F. K. & Weissleder, R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc. Natl Acad. Sci. USA 106, 12459–12464 (2009).
Wada, M. et al. Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 108, 779–788 (2010).
Tristan, A. et al. Global distribution of Panton–Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13, 594–600 (2007).
Perez, J. M., Josephson, L., O'Loughlin, T., Hogemann, D. & Weissleder, R. Magnetic relaxation switches capable of sensing molecular interactions. Nature Biotechnol. 20, 816–820 (2002).
Chung, H. J. et al. Ubiquitous detection of Gram-positive bacteria with bioorthogonal magnetofluorescent nanoparticles. ACS Nano 5, 8834–8841 (2011).
Budin, G., Chung, H. J., Lee, H. & Weissleder, R. A magnetic Gram stain for bacterial detection. Angew. Chem. Int. Ed. 51, 7752–7755 (2012).
Fraser, C. M., Eisen, J. A. & Salzberg, S. L. Microbial genome sequencing. Nature 406, 799–803 (2000).
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
Petrosino, J. F., Highlander, S., Luna, R. A., Gibbs, R. A. & Versalovic, J. Metagenomic pyrosequencing and microbial identification. Clin. Chem. 55, 856–866 (2009).
Ince, J. & McNally, A. Development of rapid, automated diagnostics for infectious disease: advances and challenges. Exp. Rev. Med. Dev. 6, 641–651 (2009).
Maurer, J. J. Rapid detection and limitations of molecular techniques. Annu. Rev. Food. Sci. Technol. 2, 259–279 (2011).
Issadore, D. et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-Hall detector. Sci. Transl. Med. 4, 141ra92 (2012).
Josephson, L., Tung, C. H., Moore, A. & Weissleder, R. High-efficiency intracellular magnetic labeling with novel superparamagnetic-Tat peptide conjugates. Bioconj. Chem. 10, 186–191 (1999).
Issadore, D. et al. Miniature magnetic resonance system for point-of-care diagnostics. Lab Chip 11, 2282–2287 (2011).
MacDougall, D. & Crummett, W. B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 52, 2242–2249 (1980).
Acknowledgements
The authors thank Y. Fisher-Jeffes for reviewing the manuscript, N. Sergeyev for synthesis of MNPs, S. Chen and M. Mckee for help with electron microscopy, C. Min for help with the µNMR device, and J. Chung, K.S. Yang, J. Carlson, H. Shao and A.V. Ullal for assistance and many helpful discussions. The work was funded in part by National Institute of Health (grants R01EB004626, R01EB010011, HHSN268201000044C and R01HL113156).
Author information
Authors and Affiliations
Contributions
H.J.C. designed and performed the research, and co-wrote the manuscript. R.W. and H.L. designed the research and wrote the manuscript. R.W. provided overall guidance. H.L. reviewed the magnetic resonance measurement data. C.M.C. provided guidance and assistance regarding the clinical studies. H.I. performed scanning electron microscopy and atomic force microscopy. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1601 kb)
Rights and permissions
About this article
Cite this article
Chung, H., Castro, C., Im, H. et al. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nature Nanotech 8, 369–375 (2013). https://doi.org/10.1038/nnano.2013.70
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2013.70
This article is cited by
-
Single-cell pathogen diagnostics for combating antibiotic resistance
Nature Reviews Methods Primers (2023)
-
Aggregation-induced emission: recent applications in infectious diseases
Science China Chemistry (2023)
-
Story of Pore-Forming Proteins from Deadly Disease-Causing Agents to Modern Applications with Evolutionary Significance
Molecular Biotechnology (2023)
-
Nanobiotics against antimicrobial resistance: harnessing the power of nanoscale materials and technologies
Journal of Nanobiotechnology (2022)
-
Emerging trends in the nanomedicine applications of functionalized magnetic nanoparticles as novel therapies for acute and chronic diseases
Journal of Nanobiotechnology (2022)