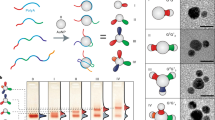
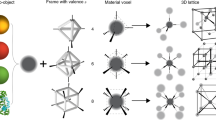
Abstract
Nanoscale components can be self-assembled into static three-dimensional structures1,2,3,4,5,6, arrays7,8,9 and clusters10,11,12,13 using biomolecular motifs. The structural plasticity of biomolecules and the reversibility of their interactions can also be used to make nanostructures that are dynamic, reconfigurable and responsive. DNA has emerged as an ideal biomolecular motif for making such nanostructures, partly because its versatile morphology permits in situ conformational changes using molecular stimuli12,14,15,16,17,18,19,20,21,22. This has allowed DNA nanostructures to exhibit reconfigurable topologies and mechanical movement17,18. Recently, researchers have begun to translate this approach to nanoparticle interfaces18,23,24, where, for example, the distances between nanoparticles can be modulated, resulting in a distance-dependent plasmonic response18,23,25. Here, we report the assembly of nanoparticles into three-dimensional superlattices and dimer clusters, using a reconfigurable DNA device that acts as an interparticle linkage. The interparticle distances in the superlattices and clusters can be modified, while preserving structural integrity, by adding molecular stimuli (simple DNA strands) after the self-assembly processes has been completed. Both systems were found to switch between two distinct rigid states, but a transition to a flexible device configuration within a superlattice showed a significant hysteresis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Hill, H. D., Macfarlane, R. J., Senesei, A. J., Lee, B., Park, S. Y. & Mirkin, C. A. Controlling the lattice parameters of gold nanoparticle FCC crystals with duplex DNA linkers. Nano Lett. 8, 2341–2344 (2008).
Maye, M. M., Nykypanchuk, D., van der Lelie, D. & Gang, O. A simple method for kinetic control of DNA-induced nanoparticle assembly. J. Am. Chem. Soc. 128, 14020–14021 (2006).
Xiong, H., van der Lelie, D. & Gang, O. Phase behavior of nanoparticles assembled by DNA linkers. Phys. Rev. Lett. 102, 015504 (2009).
Pinto, Y. Y. et al. Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett. 5, 2399–2402 (2005).
Hazarika, P., Ceyhan, B. & Niemeyer, C. M. Reversible switching of DNA–gold nanoparticle aggregation. Angew. Chem. Int. Ed. 43, 6469–6471 (2004).
Deng, Z. X., Tian, Y., Lee, S. H., Ribbe, A. E. & Mao, C. D. DNA-encoded self-assembly of gold nanoparticles into one-dimensional arrays. Angew. Chem. Int. Ed. 44, 3582–3585 (2005).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996).
Claridge, S. A., Liang, H. Y. W., Basu, S. R., Frechet, J. M. J. & Alivisatos, A. P. Isolation of discrete nanoparticle–DNA conjugates for plasmonic applications. Nano Lett. 8, 1202–1206 (2008).
Aldaye, F. A. & Sleiman, H. F. Dynamic DNA templates for discrete gold nanoparticle assemblies: control of geometry, modularity, write/erase and structural switching. J. Am. Chem. Soc. 129, 4130–4131 (2007).
Maye, M. M., Nykypanchuk, D., Cuisinier, M., van der Lelie, D. & Gang, O. Stepwise surface encoding for high-throughput assembly of nano-clusters. Nature Mater. 8, 388–391 (2009).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008).
Barish, R. D., Rothemund, P. W. K. & Winfree, E. Two computational primitives for algorithmic self-assembly: copying and counting. Nano Lett. 5, 2586–2592 (2005).
Feng, L. P., Park, S. H., Reif, J. H. & Yan, H. A two-state DNA lattice switched by DNA nanoactuator. Angew. Chem. Int. Ed. 42, 4342–4346 (2003).
Sebba, D. S., Mock, J. J., Smith, D. R., LaBean, T. H. & Lazarides, A. A. Reconfigurable core–satellite nanoassemblies as molecularly-driven plasmon switches. Nano Lett. 8, 1803–1808 (2008).
Yan, H., Zhang, X., Shen, Z. & Seeman, N. C. A robust DNA mechanical device controlled by hybridization topology. Nature 415, 62–65 (2002).
Sherman, W. B. & Seeman, N. C. A precisely controlled DNA biped walking device. Nano Lett. 4, 1203–1207 (2004).
Tian, Y. & Mao, C. D. Molecular gears: a pair of DNA circles continuously rolls against each other. J. Am. Chem. Soc. 126, 11410–11411 (2004).
Yurke, B., Turberfield, A. J., Mills, A. P., Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Sebba, D. S., LaBean, T. H. & Lazarides, A. A. Plasmon coupling in binary metal core–satellite assemblies. Appl. Phys. B 93, 69–78 (2008).
Leunissen, M. J. et al. Switchable self-protected attractions in DNA-functionalized colloids. Nature Mater. 8, 590–595 (2009).
Sonnichsen, C., Reinhard, B. M., Liphardt, J. & Alivisatos, A. P. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nature Biotechnol. 6, 741–745 (2005).
Park, S. J., Lazarides, A. A., Mirkin, C. A. & Letsinger, R. L. Directed assembly of periodic materials from protein and oligonucleotide-modified nanoparticle building blocks. Angew. Chem. Int. Ed. 40, 2909–2912 (2001).
Tkachenko, A. V. Morphological diversity of DNA–colloidal self-assembly. Phys. Rev. Lett. 89, 148303 (2002).
Valignat, M. P., Theodoly, O., Crocker, J. C., Russel, W. B. & Chaikin, P. M. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc. Natl Acad. Sci. USA 102, 4225–4229 (2005).
Mao, C. & Chen, Y. pH-induced reversible expansion/contraction of gold nanoparticle aggregates. Small 4, 2191–2194 (2008).
Park, S. J., Lazarides, A. A., Storhoff, J. J., Pesce, L. & Mirkin, C. A. The structural characterization of oligonucleotide-modified gold nanoparticle networks formed by DNA hybridization. J. Phys. Chem. B 108, 12375–12380 (2004).
Acknowledgements
This research was supported by the US Department of Energy Office of Science and Office of Basic Energy Sciences under contract no. DE-AC-02-98CH10886. The authors thank the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL) for use of their facilities. M.T.K. and W.B.S. acknowledge support by a Laboratory Directed Research and Development grant (07-025) from BNL. M.M.M. acknowledges a Goldhaber Distinguished Fellowship at BNL sponsored by Brookhaven Science Associates.
Author information
Authors and Affiliations
Contributions
M.M.M., D.N., W.B.S. and O.G. contributed to the design of the experiment and manuscript preparation. M.T.K. and W.B.S. designed and purified the DNA. M.M.M. performed synthesis and assembly experiments. M.M.M., D.N. and O.G. carried out SAXS experiments and data analysis. O.G. directed the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1221 kb)
Rights and permissions
About this article
Cite this article
Maye, M., Kumara, M., Nykypanchuk, D. et al. Switching binary states of nanoparticle superlattices and dimer clusters by DNA strands. Nature Nanotech 5, 116–120 (2010). https://doi.org/10.1038/nnano.2009.378
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2009.378
This article is cited by
-
Flexible synthesis of high-purity plasmonic assemblies
Nano Research (2021)
-
Complex assemblies and crystals guided by DNA
Nature Materials (2020)
-
DNA-Driven Nanoparticle Assemblies for Biosensing and Bioimaging
Topics in Current Chemistry (2020)
-
Crystal engineering with DNA
Nature Reviews Materials (2019)
-
Spermine induced reversible collapse of deoxyribonucleic acid-bridged nanoparticle-based assemblies
Nano Research (2018)