Abstract
Mechanisms underlying motor neuron subtype–selective endoplasmic reticulum (ER) stress and associated axonal pathology in amyotrophic lateral sclerosis (ALS) remain unclear. Here we show that the molecular environment of the ER between motor neuron subtypes is distinct, with characteristic signatures. We identify cochaperone SIL1, mutated in Marinesco-Sjögren syndrome (MSS), as being robustly expressed in disease-resistant slow motor neurons but not in ER stress–prone fast-fatigable motor neurons. In a mouse model of MSS, we demonstrate impaired ER homeostasis in motor neurons in response to loss of SIL1 function. Loss of a single functional Sil1 allele in an ALS mouse model (SOD1-G93A) enhanced ER stress and exacerbated ALS pathology. In SOD1-G93A mice, SIL1 levels were progressively and selectively reduced in vulnerable fast-fatigable motor neurons. Mechanistically, reduction in SIL1 levels was associated with lowered excitability of fast-fatigable motor neurons, further influencing expression of specific ER chaperones. Adeno-associated virus–mediated delivery of SIL1 to familial ALS motor neurons restored ER homeostasis, delayed muscle denervation and prolonged survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Saxena, S. & Caroni, P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 71, 35–48 (2011).
Cleveland, D.W. & Rothstein, J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2, 806–819 (2001).
Boillée, S., Vande Velde, C. & Cleveland, D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron 52, 39–59 (2006).
Kanning, K.C., Kaplan, A. & Henderson, C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440 (2010).
Frey, D. et al. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J. Neurosci. 20, 2534–2542 (2000).
Pun, S., Santos, A.F., Saxena, S., Xu, L. & Caroni, P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by CNTF. Nat. Neurosci. 9, 408–419 (2006).
Hegedus, J., Putman, C.T., Tyreman, N. & Gordon, T. Preferential motor unit loss in the SOD1 G93A transgenic mouse model of amyotrophic lateral sclerosis. J. Physiol. (Lond.) 586, 3337–3351 (2008).
Saxena, S., Cabuy, E. & Caroni, P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 12, 627–636 (2009).
Lee, J. & Ozcan, U. Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 289, 1203–1211 (2014).
Wang, S. & Kaufman, R.J. How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr. Opin. Lipidol. 25, 125–132 (2014).
Doyle, K.M. et al. Unfolded proteins and endoplasmic reticulum stress in neurodegenerative disorders. J. Cell. Mol. Med. 15, 2025–2039 (2011).
Hetz, C. & Mollereau, B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15, 233–249 (2014).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Wang, S. & Kaufman, R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 197, 857–867 (2012).
Kimata, Y. & Kohno, K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 23, 135–142 (2011).
Hetz, C., Martinon, F., Rodriguez, D. & Glimcher, L.H. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 91, 1219–1243 (2011).
Saxena, S. et al. Neuroprotection through excitability and mTOR required in ALS motoneurons to delay disease and extend survival. Neuron 80, 80–96 (2013).
Kaplan, A. et al. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81, 333–348 (2014).
Zhao, L., Longo-Guess, C., Harris, B.S., Lee, J.W. & Ackerman, S.L. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 37, 974–979 (2005).
Chung, K.T., Shen, Y. & Hendershot, L.M. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J. Biol. Chem. 277, 47557–47563 (2002).
Anttonen, A.K. et al. The gene disrupted in Marinesco-Sjögren syndrome encodes SIL1, an HSPA5 cochaperone. Nat. Genet. 37, 1309–1311 (2005).
Senderek, J. et al. Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat. Genet. 37, 1312–1314 (2005).
Roos, A. et al. Myopathy in Marinesco-Sjogren syndrome links endoplasmic reticulum chaperone dysfunction to nuclear envelope pathology. Acta Neuropathol. 127, 761–777 (2014).
Krieger, M. et al. SIL1 mutations and clinical spectrum in patients with Marinesco-Sjogren syndrome. Brain 136, 3634–3644 (2013).
Gorbatyuk, M.S. et al. Glucose regulated protein 78 diminishes alpha-synuclein neurotoxicity in a rat model of Parkinson disease. Mol. Ther. 20, 1327–1337 (2012).
Yan, M., Li, J. & Sha, B. Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem. J. 438, 447–455 (2011).
Nishitoh, H. et al. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 22, 1451–1464 (2008).
Bernard-Marissal, N. et al. Reduced calreticulin levels link endoplasmic reticulum stress and Fas-triggered cell death in motor neurons vulnerable to ALS. J. Neurosci. 32, 4901–4912 (2012).
Homma, K. et al. SOD1 as a molecular switch for initiating the homeostatic ER stress response under zinc deficiency. Mol. Cell 52, 75–86 (2013).
Walker, A.K. et al. ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS ONE 8, e81170 (2013).
Atkin, J.D. et al. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol. Dis. 30, 400–407 (2008).
Vucic, S. & Kiernan, M.C. Upregulation of persistent sodium conductances in familial ALS. J. Neurol. Neurosurg. Psychiatry 81, 222–227 (2010).
Turner, M.R. et al. Abnormal cortical excitability in sporadic but not homozygous D90A SOD1 ALS. J. Neurol. Neurosurg. Psychiatry 76, 1279–1285 (2005).
Wegorzewska, I., Bell, S., Cairns, N.J., Miller, T.M. & Baloh, R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA 106, 18809–18814 (2009).
Ma, H., Groth, R.D., Wheeler, D.G., Barrett, C.F. & Tsien, R.W. Excitation-transcription coupling in sympathetic neurons and the molecular mechanism of its initiation. Neurosci. Res. 70, 2–8 (2011).
Slemmer, J.E., De Zeeuw, C.I. & Weber, J.T. Don't get too excited: mechanisms of glutamate-mediated Purkinje cell death. Prog. Brain Res. 148, 367–390 (2005).
Kitao, Y. et al. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J. Neurosci. 24, 1486–1496 (2004).
Prahlad, V. & Morimoto, R.I. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 19, 52–61 (2009).
Wang, J. et al. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 5, e1000350 (2009).
Kikuchi, H. et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci. USA 103, 6025–6030 (2006).
Prause, J. et al. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 1581–1600 (2013).
Farg, M.A. et al. Mutant FUS induces endoplasmic reticulum stress in amyotrophic lateral sclerosis and interacts with protein disulfide-isomerase. Neurobiol. Aging 33, 2855–2868 (2012).
Ilieva, E.V. et al. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain 130, 3111–3123 (2007).
Uehara, T. et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441, 513–517 (2006).
Shatunov, A. et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 9, 986–994 (2010).
Watts, G.D. et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381 (2004).
Johnson, J.O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010).
Senderek, J. et al. Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am. J. Hum. Genet. 84, 511–518 (2009).
Johnson, J.O. et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17, 664–666 (2014).
Zimmer, C., Gosztonyi, G., Cervos-Navarro, J., von Moers, A. & Schroder, J.M. Neuropathy with lysosomal changes in Marinesco-Sjögren syndrome: fine structural findings in skeletal muscle and conjunctiva. Neuropediatrics 23, 329–335 (1992).
Schaeren-Wiemers, N. & Gerfin-Moser, A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431–440 (1993).
Mueller, M. et al. Macrophage response to peripheral nerve injury: the quantitative contribution of resident and hematogenous macrophages. Lab. Invest. 83, 175–185 (2003).
Rishal, I., Rozenbaum, M. & Fainzilber, M. Axoplasm isolation from rat sciatic nerve. J. Vis. Exp. 2010, e2087 (2010).
Gunasekera, K., Wuthrich, D., Braga-Lagache, S., Heller, M. & Ochsenreiter, T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13, 556 (2012).
Al Kaabi, A., Traupe, T., Stutz, M., Buchs, N. & Heller, M. Cause or effect of arteriogenesis: compositional alterations of microparticles from CAD patients undergoing external counterpulsation therapy. PLoS ONE 7, e46822 (2012).
Dirren, E. et al. Intracerebroventricular injection of adeno-associated virus 6 and 9 vectors for cell type-specific transgene expression in the spinal cord. Hum. Gene Ther. 25, 109–120 (2014).
Stevens, J.C. et al. Modification of superoxide dismutase 1 (SOD1) properties by a GFP tag–implications for research into amyotrophic lateral sclerosis (ALS). PLoS ONE 5, e9541 (2010).
Shen, J., Chen, X., Hendershot, L. & Prywes, R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111 (2002).
Gros-Louis, F., Soucy, G., Lariviere, R. & Julien, J.P. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J. Neurochem. 113, 1188–1199 (2010).
Park, K.H.J., Franciosi, S. & Leavitt, B.R. Postnatal muscle modification by myogenic factors modulates neuropathology and survival in an ALS mouse model. Nat. Commun. 4, 2906 (2013).
Acknowledgements
We thank M. Heller (Department of Clinical Research, Inselspital, Bern) for mass spectrometry, C. Müller (Pathology Institute, University of Bern) for laser microdissection microscope application, and F. Pidoux and V. Padrun (Brain Mind Institute, EPFL, Lausanne) for the generation of viruses. A.F.d.L. was supported by the International Foundation for Research in Paraplegia and the Swiss Foundation for Research on Muscle Diseases grants. N.M., C.R. and S.S. were supported by a Swiss National Science Foundation Professorship grant (PP00P3_128460) to S.S. and the Frick Foundation for ALS Research. A.G., A.R. and J.W. were supported by a START program grant, Medical Faculty, RWTH Aachen University, by the Interdisciplinary Center for Clinical Research, IZKF Aachen (N5–3), and by the German Research Foundation, DFG (WE 1406/13–1).
Author information
Authors and Affiliations
Contributions
S.S. and A.F.d.L. conceived the study, wrote the manuscript and performed the experiments with contributions from N.M. (biochemistry), M.C.B. (cell culture, qPCR analyses), C.R. (cloning of AAV vectors) and R.R. (mouse breeding, behavior experiments and tissue collection). B.L.S. generated viruses and gave advice for viral experiments. D.T. examined ALS and control autopsies and provided diagnoses. A.G. performed the immunohistochemical analyses of the human autopsy samples. A.R. and J.W. performed electron microscopic examinations. All authors read and commented on the manuscript. S.S. supervised the overall project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
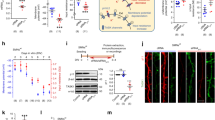
Supplementary Figure 1 Genetically reducing SIL1 expression enhances ER stress responses in motor neurons.
(a) Immunolabeling of motor neurons for ChAT and BiP reveals high BiP levels in Sil1+/– (yellow arrow) and Sil1−/− motor neurons (MNs). WT motor neurons showing low levels of BiP (white arrows). Right: Scatter plot, labeling intensities plotted as arbitrary units (a.u.), shows BiP levels in ChAT positive motor neurons WT (n = 195), Sil1+/– (n = 145) and Sil1−/− (n = 147 motor neurons from 4 mice / genotype), (one-way ANOVA, F2, 384 = 322.9, *** P < 0.0001). Lower box and whiskers plot: % of motor neurons with high BiP levels in Sil1+/–, n = 8, Sil1−/−, n = 13 and control WT, n = 10 sections / mice from 4 mice / genotype, (one-way ANOVA, F2, 186 = 28304, *** P < 0.0001).
(b) Immunolabeling of Sil1−/− spinal cord with Neuronal marker NeuN and Pi-eIF2α, wide spread UPR in spinal neurons besides motor neurons. Note the striking absence of Pi-eIF2α immunoreactivity in the dorsal horn neurons (arrow).
(c) Pi-eIF2α positive cerebellar PCs are present in Sil1−/− condition (yellow arrows). Note the prominent atrophy of the cell soma and dendrites. Right: % of Pi-eIF2α positive PCs and motor neurons in WT, Sil1+/– and Sil1−/−, (WT, n = 10, Sil1+/–, n = 6 and Sil1−/−, n = 12 sections / mice from 4 mice each genotype, (Unpaired t-test, t = 14.18, *** P < 0.0001).
(d) Immunoblotting of muscles indicates mild UPR in Sil1+/– and strong UPR in Sil1−/− mice at 19 weeks of age, n = 3 mice / genotype, (one-way ANOVA, F3, 56 = 22805, *** P < 0.0001). RB out of three experiments. Scale: (a, c) 15 and (b) 50 µm.
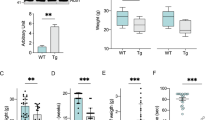
Supplementary Figure 2 Specific alterations in the spinal cord and NMJs in woozy (Sil1−/−) mice.
(a) NMJs labeled with neurofilament (NF-200) and α-Bungarotoxin (α-BTX) reveals partially occupied NMJs only in Sil1−/− (blue arrows) but not in Sil1+/– mice (yellow arrows, normal innervation). Right: Representative images showing criteria for assessment of axonal innervation and % of NMJs classified according to the nerve occupancy at the end plates: more than 80% (fully innervated), more than 30% (partially innervated) and less than 30% (denervated).
(b) WT littermate with normal esterase staining pattern. Right: Grouped partially atrophic and angular muscle fibers (arrows) indicating neurogenic atrophy in Sil1−/− or woozy (wz) mouse muscle n = 6 mice each genotype.
(c) Electron microscopy reveals autophagic material in axon terminal (arrow) at the end plate. Note the lobulated shape of the synapse-associated muscle fiber nuclei typical for the wz phenotype, n = 12 mice.
(d) Autophagic material (arrows) in axons of terminal myelinated intramuscular nerve fibers of wz mouse.
(e) Microglia activation as reported in the cerebellum is present in the spinal cord, revealed by immunostaining for a microglia marker CD11b. Right: Box and whiskers plot depicting % of CD11b positive area in the grey matter of spinal cord, (WT, n = 8, Sil1+/–, n = 5 and Sil1−/−, n = 11 sections / mice from 4 mice each genotype, one-way ANOVA, F3, 31 = 15.03, *** P < 0.0001). Scale: (a) 10 and 5 (b) 80, (c) 2.5, (d) 1 and (e) 30 μm.
Supplementary Figure 3 Genetic ablation of SIL1 affects axonal regenerative responses.
(a) Venn diagram of proteins detected in the axoplasm at basal level in non-lesioned WT and Sil1+/– sciatic nerve using mass spectrometry.
(b) Pie chart indicating the percentage of proteins altered in expression 24 h after sciatic nerve lesion in Sil1+/– and in WT axoplasm. Comparison was only made for proteins detected at basal level in both genotypes. Cut off parameters used (PMSS score greater than 40, unique peptides detected more than 4), n = 8 mice per genotype for each condition.
(c) Proteins were plotted using semi-quantitative PMSS score. Several proteins were upregulated in the axoplasm in response to nerve lesion in WT condition but remained unchanged in Sil1+/– lesioned axoplasm. Mtap1b and Tub2a are examples of proteins which are upregulated in response to nerve lesion in both genotypes.
(d) Proteins downregulated in Sil1+/– axoplasm 24 h after nerve crush compared to control non-lesioned side were grouped into protein classes according to PANTHER analyses.
Supplementary Figure 4 Specificity of SIL1 antibody in spinal motor neurons.
50 μm spinal cord sections were immunostained for SIL1 and BiP. Note the reduction in SIL1 labeling in Sil1+/– motor neurons compared to WT motor neurons and the complete loss of SIL1 labeling in Sil1−/− spinal motor neurons. Increase in BiP levels parallels the decline in SIL1 levels. Scale: 15 μm.
Supplementary Figure 5 SIL1 levels are selectively reduced in spinal motor neurons in fALS mouse models.
(a) SIL1 immunolabeling is diminished in motor neurons in G93A-s spinal cord whereas the dorsal horn displays SIL1 positive cells. Right: Immunolabeling indicates the presence of B8H10 misfSOD1 epitope only in VAChT positive motor neurons.
(b) Loss of SIL1 in a sub-population of motor neurons precedes UPR signaling. Motor neurons lacking SIL1 show Pi-eIF2α expression at P140 in G93A-s mice (yellow arrows). % of motor neurons (MNs) expressing Pi-eIF2α or SIL1 over age in disease, n = 3 mice / genotype.
(c) Immunoblotting of cerebellar protein lysates and immunostaining of cerebellum for SIL1 in G93A-s mice indicates no alterations in SIL1 levels compared to WT condition. n = 3 mice each age, RB out of three blots. Scale: (a) 50, (b) 100 and (c) 10 μm.
Supplementary Figure 6 UPR signaling and misfSOD1 are detected in non-motor neurons in G93A-s;Sil1+/– but not in G93A-s spinal cord.
(a) Immunostaining with B8H10 antibody shows presence of misfSOD1 in ChAT negative neurons at P90, G93A-s-Sil1+/– mice, which is not detected in G93A-s condition, n = 4 mice each.
(b) Immunolabeling reveals ChAT negative neurons positive for Pi-eIF2α in P90, G93A-s-Sil1+/– but absent in P110, G93A-s mice (earliest age, UPR starts in FF motor neurons), n = 4 mice each. Scale: (a, b) 50 and zoom 15 µm.
Supplementary Figure 7 Overexpression of SIL1 leads to reduced detection of B8H10-positive SOD1-G93A and SOD1-G85R labeling.
(a) Endogenous low expression of SIL1 in n2a cells. Cells were transfected with hSIL1-myc and immunolabeled for SIL1. Compare SIL1 expression levels in transfected cells (yellow arrows) versus endogenous levels in non-transfected cells (white arrows).
(b) Schematic for sequential expression and quantification of B8H10 positive misfSOD1 intensity in transfected n2a cells (%).
(c) Cells transfected with hSIL1-myc and SOD1-G85R were fixed 24 h or 48 h after transfection. Immunolabeling displays reduced detection of B8H10 positive misfSOD1 in double transfected cells (yellow arrows) compared to SOD1-G85R only (red arrows) expressing cells. Right: quantification of B8H10 positive SOD1-G85R intensity (%). Scale: (a) 8, (c) 20 μm.
Supplementary Figure 8 Overexpression of GFP does not alter misfSOD1 levels in n2a cells, and higher SIL1 or BiP levels do not affect GFP levels.
n2a cells double transfected with GFP and SOD1-G93A show no effect of GFP expression on B8H10 positive misfSOD1 species (upper panel). Middle panel: labeling for hSIL1-myc and GFP reveal no effect on total GFP levels in SIL1 overexpressing cells. Lower panel: Overexpression of hBiP-myc does not alter GFP levels in cells. Scale: 20 μm.
Supplementary Figure 9 Identification of critical residues for SIL1-BiP interaction.
(a) Crystal structure of the yeast SIL1-BiP complex (PDB 3QML). Identification of critical SIL1 residues involved in BiP binding by the analysis of the yeast (S. cerevisiae) SIL1-BiP complex crystal structure.
(b) Clustal Omega HHPRED (http://toolkit.tuebingen.mpg.de/hhpred) sequence alignment of human (Hs) and yeast (Sc) SIL1. Critical residues are boxed in grey and indicate the position of the residues important for BiP binding in yeast. These residues were investigated by point mutations introduced in HsSIL1 in this study.
(c) Double transfection of n2a cells with point mutants: hSIL1-H214A, hSIL1-E441A and hSIL1-K288A together with SOD1-G93A, immunostaining for misfSOD1 and pan-SOD1 identifies residue H214 in HsSIL1 as critical for SIL1-BiP interaction. Mutation in this residue leads to loss of SIL1 mediated reduction in B8H10 positive misfSOD1. A minor effect on B8H10 positive misfSOD1 is also observed in the case of residue E441. Right: quantification of B8H10 positive misfSOD1 intensity in double transfected cells (%). Scale: (c) 20 μm.
Supplementary Figure 10 Motor neuron transduction profile after AAV6-SIL1-myc delivery to G93A-s mice.
(a) Schematic depicting intracerebroventricular (i.c.v.) injection of AAV6-SIL1-myc in G93A-s neonates between days P1–P2.
(b) Double immunostaining of AAV6-SIL1-myc injected spinal cord against SIL1 and myc-tag reveals strong and overlapping expression between endogenous and virally applied SIL1. Note the lack of myc-tag immunolabeling in G93A-s motor neurons.
(c) Immunolabeling against myc-tag detects AAV6-mediated SIL1 expression in ChAT positive motor neurons in adult G93A-s mice. White arrows point to non-infected motor neurons whereas yellow arrows mark motor neurons presenting robust expression of AAV6-SIL1-myc. Longitudinal transduction profile across lumbar spinal cord expressed as myc+ motor neurons (MNs) against the total number of motor neurons per section as determined by ChAT staining. Scale: (b) 20, (c) 40 μm.
Supplementary Figure 11 SIL1 overexpression in motor neurons ameliorates the ALS phenotype.
(a) Representative image showing increased immunoreactivity for Derlin-1 and CRT in AAV6-SIL1 transduced motor neurons (yellow arrows) versus non-transduced motor neurons (white arrows) in G93A-f spinal cord.
(b) Representative image displaying reduced B8H10 immunopositive misfSOD1 in AAV6-SIL1 transduced G93A-f motor neurons without change in total hSOD1 levels as identified by human SOD1 antibody. Scale: (a) 100, (b) 25 μm.
(c) SIL1 overexpression in G93A-s spinal motor neurons, improved motor coordination, exercise capacity as measured by rotarod. Rotarod performance plotted in seconds as latency to fall. Error bars are mean ± s.e.m from 8 mice / genotype (Unpaired t-test, t = 5.222, *** P < 0.0001). Survival: Mean survival values were G93A-s + vehicle = 279 days, G93A-s-AAV6-SIL1 = 363 days, G93A-s-AAV6-GFP = 270 days. Log-rank (Mantel Cox-test) = 24.63, P < 0.0001. Mean survival values for G93A-f were G93A-f + vehicle = 153 days, G93A-f-AAV6-SIL1 = 190 days. Log-rank (Mantel Cox-test) = 5.629, P = 0.0177. Number of mice G93A-s + vehicle = 18 (7 males, 11 females), G93A-s-AAV6-Sil1 = 14 (7 males, 7 females), G93A-s-AAV6-GFP = 5 males, G93A-f + vehicle = 10 (6 males, 4 females) and G93A-f-AAV6-Sil1 = 6 (5 males, 1 female).
Supplementary Figure 12 SIL1 levels are not dependent on ER stress and regulate ER homeostasis.
(a) qPCR for hSOD1 mRNA from G93A-s FF, SOL motor neurons (MNs), cerebellar PCs and MC (primary motor cortex layer V), indicates equal expression of the SOD1-G93A transgene. Note the higher expression of the transgene in cerebellar PCs versus FF MNs (Unpaired t-test, t = 3.69, * P = 0.031), n = 6 mice each.
(b) Sil1 transcripts remain unchanged in motor neurons after pharmacological induction of ER stress. Note the strong increase in Bip transcripts after the induction of ER stress in motor neurons.
(c) qPCR reveals the selective downregulation of Crt transcripts in FF motor neurons but not in SOL motor neurons in fALS models. Sil1 mRNA decline precedes those of Crt.
(d) CRT levels are reduced in Sil1−/− mice. Immunostaining for CRT in Sil1−/− spinal cord and cerebellum presents a decline in expression of CRT in ChAT positive motor neurons and Calbindin positive PCs.
(e) Immunostaining for SIL1 and CRT displays no change in CRT expression after AMPA treatment in Sil1−/− mice. Right: qPCR analyses of SOL motor neurons from Sil1−/− mice show reduced mRNA for ER chaperones Erp29, Crt and Rbx1 and an increase in stress sensors Ire1 and Perk. AMPA treatment does not change this observation. Scale (d, e) 50 and PCs 10 μm.
Supplementary Figure 13 Model of how motor neuron subtype-selective expression of SIL1 influences disease manifestations.
(a) The model has two main implications about the subtype-specific manifestation of ER stress in SOD1-G93A models. First, low excitable FF motor neurons (MNs) intrinsically express low levels of SIL1 in comparison to disease-resistant S motor neurons (shadings of light to dark indicate low to high expression of a particular protein between figure panels a and b). In mutant FF motor neurons an unidentified specific cellular stressor impairs Ca2+ homeostasis, altering motor neuron excitability, and further reduces SIL1 levels. Reduced SIL1 levels negatively impacts SIL1-BiP coupling and compromises ER homeostasis. Imbalance in ER homeostasis downregulates the expression of critical chaperones such as CRT, ERP29 and RBX1, leading to reduced chaperone activity and altered ER homeostasis. This in turn probably generates a feedback loop further impacting Ca2+ homeostasis and particularly sensitizing FF motor neurons. Secondly, inefficient nucleotide exchange of BiP overtime leads to increased expression and accumulation of substrate bound BiP-ADP, initiating UPR in FF motor neurons, followed by axonal denervation.
(b) Disease resistant highly excitable S motor neurons intrinsically express high levels of SIL1 which promotes efficient nucleotide exchange of BiP and sustained ER homeostasis. To cope with increasing age associated disease burden, S motor neurons further upregulate SIL1 expression. Hypothetical or unknown mechanisms are suggested by dashed arrows with question marks.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Table 1 (PDF 11502 kb)
Source data
Rights and permissions
About this article
Cite this article
Filézac de L'Etang, A., Maharjan, N., Cordeiro Braña, M. et al. Marinesco-Sjögren syndrome protein SIL1 regulates motor neuron subtype-selective ER stress in ALS. Nat Neurosci 18, 227–238 (2015). https://doi.org/10.1038/nn.3903
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3903
This article is cited by
-
Selective vulnerability of motor neuron types and functional groups to degeneration in amyotrophic lateral sclerosis: review of the neurobiological mechanisms and functional correlates
Brain Structure and Function (2023)
-
Heat shock protein Grp78/BiP/HspA5 binds directly to TDP-43 and mitigates toxicity associated with disease pathology
Scientific Reports (2022)
-
PolyGA targets the ER stress-adaptive response by impairing GRP75 function at the MAM in C9ORF72-ALS/FTD
Acta Neuropathologica (2022)
-
Pathomechanisms of ALS8: altered autophagy and defective RNA binding protein (RBP) homeostasis due to the VAPB P56S mutation
Cell Death & Disease (2021)
-
Endoplasmic reticulum and mitochondria in diseases of motor and sensory neurons: a broken relationship?
Cell Death & Disease (2018)