Abstract
DNA damage is considered to be a prime factor in several spinocerebellar neurodegenerative diseases; however, the DNA lesions underpinning disease etiology are unknown. We observed the endogenous accumulation of pathogenic topoisomerase-1 (Top1)-DNA cleavage complexes (Top1ccs) in murine models of ataxia telangiectasia and spinocerebellar ataxia with axonal neuropathy 1. We found that the defective DNA damage response factors in these two diseases cooperatively modulated Top1cc turnover in a non-epistatic and ATM kinase–independent manner. Furthermore, coincident neural inactivation of ATM and DNA single-strand break repair factors, including tyrosyl-DNA phosphodiesterase-1 or XRCC1, resulted in increased Top1cc formation and excessive DNA damage and neurodevelopmental defects. Notably, direct Top1 poisoning to elevate Top1cc levels phenocopied the neuropathology of the mouse models described above. Our results identify a critical endogenous pathogenic lesion associated with neurodegenerative syndromes arising from DNA repair deficiency, indicating that genome integrity is important for preventing disease in the nervous system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
McKinnon, P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 10, 100–112 (2009).
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
O'Driscoll, M. & Jeggo, P.A. The role of double-strand break repair—insights from human genetics. Nat. Rev. Genet. 7, 45–54 (2006).
McKinnon, P.J. Maintaining genome stability in the nervous system. Nat. Neurosci. 16, 1523–1529 (2013).
Shull, E.R. et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 23, 171–180 (2009).
Suberbielle, E. et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat. Neurosci. 16, 613–621 (2013).
Lu, T. et al. Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004).
Lavin, M.F. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signaling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769 (2008).
Lee, Y. & McKinnon, P.J. Responding to DNA double strand breaks in the nervous system. Neuroscience 145, 1365–1374 (2007).
Shiloh, Y. & Ziv, Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013).
Stracker, T.H. & Petrini, J.H. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011).
Taylor, A.M., Groom, A. & Byrd, P.J. Ataxia-telangiectasia-like disorder (ATLD): its clinical presentation and molecular basis. DNA Repair (Amst.) 3, 1219–1225 (2004).
Takai, H. et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21, 5195–5205 (2002).
Date, H. et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 29, 184–188 (2001).
Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 (2008).
McKinnon, P.J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 7, 303–321 (2012).
Takashima, H. et al. Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272 (2002).
Ahel, I. et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443, 713–716 (2006).
El-Khamisy, S.F. et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005).
Alagoz, M., Chiang, S.C., Sharma, A. & El-Khamisy, S.F. ATM deficiency results in accumulation of DNA–topoisomerase I covalent intermediates in neural cells. PLoS ONE 8, e58239 (2013).
Lin, C.P., Ban, Y., Lyu, Y.L., Desai, S.D. & Liu, L.F. A ubiquitin-proteasome pathway for the repair of topoisomerase I–DNA covalent complexes. J. Biol. Chem. 283, 21074–21083 (2008).
Sordet, O. et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I–induced DNA double-strand breaks. EMBO Rep. 10, 887–893 (2009).
Pommier, Y. et al. Repair of topoisomerase I–mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81, 179–229 (2006).
Wang, J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002).
Wu, H.Y. & Liu, L.F. DNA looping alters local DNA conformation during transcription. J. Mol. Biol. 219, 615–622 (1991).
Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Katyal, S. et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 26, 4720–4731 (2007).
Nitiss, J.L., Soans, E., Rogojina, A., Seth, A. & Mishina, M. Topoisomerase Assays (John Wiley & Sons, 2012).
Subramanian, D., Rosenstein, B.S. & Muller, M.T. Ultraviolet-induced DNA damage stimulates topoisomerase I–DNA complex formation in vivo: possible relationship with DNA repair. Cancer Res. 58, 976–984 (1998).
Heideker, J., Prudden, J., Perry, J.J., Tainer, J.A. & Boddy, M.N. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1-mediated DNA damage and genome instability. PLoS Genet. 7, e1001320 (2011).
Hsiang, Y.H., Hertzberg, R., Hecht, S. & Liu, L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260, 14873–14878 (1985).
Sakasai, R., Teraoka, H., Takagi, M. & Tibbetts, R.S. Transcription-dependent activation of ataxia telangiectasia mutated prevents DNA-dependent protein kinase–mediated cell death in response to topoisomerase I poison. J. Biol. Chem. 285, 15201–15208 (2010).
Chiang, S.C., Carroll, J. & El-Khamisy, S.F. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle 9, 588–595 (2010).
Das, B.B. et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 28, 3667–3680 (2009).
Hickson, I. et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64, 9152–9159 (2004).
Lin, C.P., Ban, Y., Lyu, Y.L. & Liu, L.F. Proteasome-dependent processing of topoisomerase I–DNA adducts into DNA double strand breaks at arrested replication forks. J. Biol. Chem. 284, 28084–28092 (2009).
Mao, Y., Sun, M., Desai, S.D. & Liu, L.F. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA 97, 4046–4051 (2000).
Herzog, K.H., Chong, M.J., Kapsetaki, M., Morgan, J.I. & McKinnon, P.J. Requirement for Atm in ionizing radiation–induced cell death in the developing central nervous system. Science 280, 1089–1091 (1998).
Lee, Y. et al. ATR maintains select progenitors during nervous system development. EMBO J. 31, 1177–1189 (2012).
Lee, Y. et al. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat. Neurosci. 12, 973–980 (2009).
Pourquier, P. et al. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J. Biol. Chem. 272, 26441–26447 (1997).
Pourquier, P. et al. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J. Biol. Chem. 272, 7792–7796 (1997).
Beal, M.F. Oxidatively modified proteins in aging and disease. Free Radic. Biol. Med. 32, 797–803 (2002).
Palmeri, S. et al. Clinical course of two Italian siblings with ataxia-telangiectasia-like disorder. Cerebellum 12, 596–599 (2013).
Vos, S.M., Tretter, E.M., Schmidt, B.H. & Berger, J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 (2011).
Iossifov, I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
Neale, B.M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012).
King, I.F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013).
Stoll, G. et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237 (2013).
Xu, D. et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 16, 1238–1247 (2013).
Acknowledgements
We thank E. Soans and M. Mishina for assistance with the ICE bioassay, B. Kuzio for general technical assistance, F. Alt (Children's Hospital of Boston) for Prkdc−/− mice, K. Caldecott and S. El-Khamisy (U. Sussex) and R. Klein-Geltink (St. Jude Children's Research Hospital) for helpful discussions and S. Foster (Memorial Sloan-Kettering Cancer Center) for help analyzing the mice. We also thank the St. Jude Children's Research Hospital Animal Resource Center and the Transgenic Core Unit for support with mouse work. P.J.M. is supported by the US National Institutes of Health (NS-37956, CA-96832), the CCSG (P30 CA21765), and the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital. J.L.N. is supported by the National Cancer Institute (CA52814 and CA82313). J.H.J.P. is supported by the US National Institutes of Health (GM59413), the Geoffrey Beene Foundation and the Goodwin Foundation. Y. Lee is supported by the SRC program (2011-0030833). S.K. is a Neoma Boadway AP Endowed Fellow and is supported by grants from the University of Manitoba, CancerCare Manitoba and a Manitoba Health Research Council Establishment award.
Author information
Authors and Affiliations
Contributions
S.K. and P.J.M. conceived and planned all of the experiments and produced the final version of the manuscript. S.K. performed all of the experiments with contributions from K.C.N. (in vitro TDP1 cleavage assay), Y. Lee, M.S. and H.R.R. (generation of the mutant mice and additional technical support), S.M.D. and Y. Li (processing tissue for ICE bioassay and mouse colony management), and J.Z. (AtmNes-cre and ATMi immunoblotting experiments). J.H.J.P. contributed critical reagents and experimental results. J.L.N. contributed to experimental design and the interpretation of results and the preparation of the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 DNA strand breaks accumulate in transcriptionally-active quiescent/post-mitotic cells.
Control, Atm-/- and Tdp1-/- primary astrocyte lines fail to accumulate Top1-associated DNA breaks in the presence of 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole [DRB], a potent RNA polymerase II inhibitor. Representative scatterplots indicate cellular comet tail moments after CPT (inset scattergram panels). For each in vitro comet assay, 100 cells/comet corresponding to each genotype and treatment were analyzed and experiments were performed in triplicate (total of n=300 cells/genotype/treatment). Data from alkaline comet analysis is presented: representative scatterplots indicate cellular comet tail moments. */** denotes p-values < 0.0001.
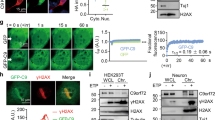
Supplementary Figure 2 Tdp1-dependent Top1-DNA cleavage activity is ATM-independent.
a. Structures of the radiolabelled phosphotyrosyl-containing substrate and the resulting cleaved product. The phosphotyrosyl bond is indicated (red arrow). b. Immunoblots of Ctrl and Atm-/- cerebella extract used in TDP1 enzymatic cleavage assay. c. Electrophoretogram of separated enzymatic products by polyacrylamide gel electrophoresis. Unprocessed radiolabelled substrate migrates at a higher molecular weight than the cleaved product. d. Quantification of TDP1 cleavage assay data from the eletrophoretogram. Control and Atm-deficient cerebellar extracts have similar TDP1 cleavage activities. Results are an average of four independent experiments. Full-length gel images are presented in Supplementary Figure 12.
Supplementary Figure 3 Top1-dependent DNA strand breaks accumulate in response to oxidative stress.
CPT-induced DNA strand breaks that accumulate in Atm-/- primary astrocyte lines show ~1/3 reduced DNA damage upon co-treatment with the anti-oxidant N-acetylcysteine (NAC). For each in vitro comet assay, 100 cells/comet corresponding to each genotype and treatment were analyzed and experiments were performed in triplicate (total of n=300 cells/genotype/treatment). * denotes p-values < 0.0001.
Supplementary Figure 4 Top1cc resolution and downregulation is ATM-dependent.
a. Analysis of total Top1 protein levels after persistent CPT treatment (14 μM, 1 and 3 hrs at 37°C). Top1 expression in control LCLs is reduced with prolonged exposure to CPT, reflecting Top1 degradation. In contrast, ~4-fold higher levels of Top1 persist with CPT treatment in ATM-/- cells. Inhibition of ATM kinase via KU55933 (10μM) co-treatment shows similar Top1 degradation as CPT-treated controls. Top1 protein levels persist after transcriptional inhibition using DRB, indicating that Top1 turnover is linked to transcription. Relative levels of Top1 are quantified respective to each mock-treated counterpart. b. ICE analysis of ATM-/- lymphoid cells following CPT treatment (14 μM, 1 and 3 hrs at 37°C) show accumulation of Top1cc compared to control counterparts. ICE blots were subsequently probed with 32P-labeled human genomic DNA (gDNA) to control for relative DNA loading. c. ICE analysis of ATM-/- lymphoid cells following CPT (14 μM, 1 hour at 37°C) and KU55933 (10μM) co-treatment indicates Top1cc accumulation is not dependent on ATM kinase function as shown by comparable levels of Top1cc in CPT-treated ctrl cells with and without KU55933 co-treatment. ICE blots were subsequently probed with 32P-labeled human genomic DNA (gDNA) to control for relative DNA loading. Full-length blots/gels are presented in Supplementary Figure 12.
Supplementary Figure 5 Top1cc poly-ubiquitination and sumoylation are regulated by ATM.
TDP1 overexpression (flag-TDP1) in HEK293T cells increases the apparent level of immunoprecipitated poly-sumoylated Top1 in both control and shATM knock-down lines (red bracket) when compared to equivalent lines expressing normal or reduced levels of TDP1. 293T clones were obtained by puromycin selection after Fugene 6 (Roche)-mediated transfection of shATM or control (shScm). pCMV-Flag vector alone, pCMV-Flag-TDP1 or shTDP1 were subsequently transfected into shATM or shScm puromycin resistant 293T cells. Full-length Western blot images are presented in Supplementary Figure 12.
Supplementary Figure 6 Atm mediates γH2AX foci formation upon Top1-DNA damage in quiescent primary fibroblasts.
a. Human fibroblasts (HFs) and b. mouse embryonic fibroblasts (MEFs) were subjected to ionizing radiation (2Gy IR with 60mins recovery at 37°C) or camptothecin (5μM for 60mins at 37°C), paraformaldehyde-fixed, immunostained with anti-γH2AX antibody followed by Alexa-555-conjugated secondary antibody. Cells were counterstained with DAPI and Alexa-488-conjugated phalloidin to indicate actin fibers and cell size. c. CPT-treatment of fibroblasts (A-T and Atm-/-) results in few γH2AX foci while radiation treatment results in a similar amount of γH2AX foci amongst ctrl, Atm-/-, Tdp1-/- and NBS-/- fibroblasts, indicating that Top1-dependent damage signaling requires functional ATM. Tdp1-/- cells form higher levels of Top1-induced γH2AX foci due to defective Top1cc processing activity. For all foci quantification experiments, 30 cells for each cell line and corresponding treatment were counted and experiments were repeated in quadruplicate (total n=120 independent cells measured per line/treatment). Bar graphs represent mean cellular foci values of all replicates, error bars represent standard error of means (S.E.M.).
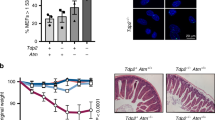
Supplementary Figure 7 Atm and Tdp1 are required for embryonal brain development.
a. Immunoanalysis of the E14.5 embryonal Atm-/-Tdp1-/- midbrain reveals an accumulation of unrepaired DNA breaks. b. These regions correlate with an increase in anti-p53 immunostaining and c. neuronal apoptosis in Tuj1-negative (proliferative) regions. Similarly, immunoanalysis of the E14.5 embryonal Atm-/-Tdp1-/- cerebellar rhombic lip and external granule layers shows an accumulation of unrepaired DNA breaks (d. and g.), increased anti-p53 immunostaining (e. and h.) and neuronal apoptosis (f. and i.).
Supplementary Figure 8 Analysis of the AtmNes-cre;Tdp1-/- developing brain.
Like the germline Atm-/-;Tdp1-/- embryos, immunoanalysis of the E14.5 embryonal AtmNes-creTdp1-/- forebrain and midbrain reveals an accumulation of unrepaired DNA breaks (yellow arrowheads, a. and d.). These regions correlate with an increase in anti-p53 immunostaining (red arrowheads, b. and e.) and neuronal apoptosis (yellow arrowheads, c. and f.). g. Immunoblot analysis showing specific deletion of Atm within the CNS in AtmNes-Cre mice (3 weeks old). Note that Atm protein is absent only in cerebellar tissue derived from AtmNes-Cre mice but remains in non-nestin-lineage tissue, such as thymus. Like Atm-/- tissue, AtmNes-Cre CNS tissue do not undergo DNA damage-induced (IR) phosphorylation of KAP1. Full-length Western blot images are presented in Supplementary Figure 12.
Supplementary Figure 9 Topotecan treatment causes apoptosis in neural tissue.
E12.5 embryos were exposed to Topotecan (0.5 μg/g body weight; +/-TPT) and then collected at E14.5 to determine the effect of Top1cc on overall embryonic development. Notably, as determined using TUNEL staining, the TPT caused apoptosis exclusively throughout the developing nervous system. Ctx is neocortical region, V is the ventricle, Thal is thalamus and Olf is the olfactory bulb region.
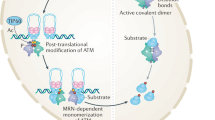
Supplementary Figure 10 Top1cc as an etiological lesion in ataxia telangiectasia.
ATM prevents Top1cc accumulation, which would otherwise result in increased strand breaks. In the absence of ATM, Top1cc induced SSBs can be converted to DSBs (during replication associated with neurogenesis) that activate MRN to promote canonical ATM kinase signaling, which can lead to apoptosis of DNA damaged cells. In non-cycling neural cells, Top1cc accumulation can result in DNA breaks that may disrupt transcription. Collectively, this scheme predicts loss of ATM both during neurogenesis and in the mature nervous system can lead to the accumulation of damaged cells in the nervous system, which eventually results in cell death and neurodegeneration.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Table 1 (PDF 9489 kb)
Rights and permissions
About this article
Cite this article
Katyal, S., Lee, Y., Nitiss, K. et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci 17, 813–821 (2014). https://doi.org/10.1038/nn.3715
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3715
This article is cited by
-
PSIP1/LEDGF reduces R-loops at transcription sites to maintain genome integrity
Nature Communications (2024)
-
Redox dysregulation as a driver for DNA damage and its relationship to neurodegenerative diseases
Translational Neurodegeneration (2023)
-
TDP1 suppresses chromosomal translocations and cell death induced by abortive TOP1 activity during gene transcription
Nature Communications (2023)
-
The threat of programmed DNA damage to neuronal genome integrity and plasticity
Nature Genetics (2022)
-
Defective repair of topoisomerase I induced chromosomal damage in Huntington’s disease
Cellular and Molecular Life Sciences (2022)