Abstract
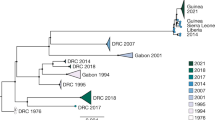
Since 2013, West Africa has encountered the largest Ebola virus (EBOV) disease outbreak on record, and Sierra Leone is the worst-affected country, with nearly half of the infections. By means of next-generation sequencing and phylogeographic analysis, the epidemiology and transmission of EBOV have been well elucidated. However, the intra-host dynamics that mainly reflect viral–host interactions still need to be studied. Here, we show a total of 710 intra-host single nucleotide variations (iSNVs) from deep-sequenced samples from EBOV-infected patients, through a well-tailored bioinformatics pipeline. We present a comprehensive distribution of iSNVs during this outbreak and along the EBOV genome. Analyses of iSNV and its allele frequency reveal that VP40 is the most conserved gene during this outbreak, and thus it would be an ideal therapeutic target. In the co-occurring iSNV network, varied iSNV sites present different selection features. Intriguingly, the T-to-C substitutions at the 3′-UTR of the nucleoprotein (NP; positions 3008 and 3011), observed in many patients, result in the upregulation of the transcription of NP through an Ebola mini-genome reporting system. Additionally, no iSNV enrichment within B-cell epitopes of GP has been observed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ebola Situation Reports WHO (2015); http://apps.who.int/ebola/current-situation/
Lyon, G. M. et al. Clinical care of two patients with Ebola virus disease in the United States. New Engl. J. Med. 371, 2402–2409 (2014).
Parra, J. M., Salmeron, O. J. & Velasco, M. The first case of Ebola virus disease acquired outside Africa. New Engl. J. Med. 371, 2439–2440 (2014).
Varkey, J. B. et al. Persistence of Ebola virus in ocular fluid during convalescence. New Engl. J. Med. 372, 2423–2427 (2015).
Deen, G. F. et al. Ebola RNA persistence in semen of Ebola virus disease survivors—preliminary report. New Engl. J. Med. http://dx.doi.org/10.1056/NEJMoa1511410 (2015).
Mate, S. E. et al. Molecular evidence of sexual transmission of Ebola virus. New Engl. J. Med. 373, 2448–2454 (2015).
Gire, S. K. et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 (2014).
Tong, Y. G. et al. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature 524, 93–96 (2015).
Carroll, M. W. et al. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature 524, 97–101 (2015).
Simon-Loriere, E. et al. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature 524, 102–104 (2015).
Park, D. J. et al. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell 161, 1516–1526 (2015).
Domingo, E., Sheldon, J. & Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76, 159–216 (2012).
Acevedo, A., Brodsky, L. & Andino, R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505, 686–690 (2014).
Li, Y. et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 42, 969–972 (2010).
Boyko, A. R. et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4, e1000083 (2008).
Emmett, K. J., Lee, A., Khiabanian, H. & Rabadan, R. High-resolution genomic surveillance of 2014 Ebolavirus using shared subclonal variants. PLoS Curr. Outbreak http://dx.doi.org/10.1371/currents.outbreaks.c7fd7946ba606c982668a96bcba43c90 (2015).
Liang, B. et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015).
Pickett, B. E. et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40, D593–D598 (2012).
Andino, R. & Domingo, E. Viral quasispecies. Virology 479–480, 46–51 (2015).
Josefsson, L. et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl Acad. Sci. USA 110, E4987–E4996 (2013).
Sobesky, R. et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology 46, 1704–1712 (2007).
Del Portillo, A. et al. Multiploid inheritance of HIV-1 during cell-to-cell infection. J. Virol. 85, 7169–7176 (2011).
Jung, A. et al. Recombination: multiply infected spleen cells in HIV patients. Nature 418, 144 (2002).
McWilliam Leitch, E. C. & McLauchlan, J. Determining the cellular diversity of hepatitis C virus quasispecies by single-cell viral sequencing. J. Virol. 87, 12648–12655 (2013).
Bornholdt, Z. A. et al. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154, 763–774 (2013).
Adu-Gyamfi, E. et al. Host cell plasma membrane phosphatidylserine regulates the assembly and budding of Ebola virus. J. Virol. 89, 9440–9453 (2015).
Kouznetsova, J. et al. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg. Microbe. Infect. 3, e84 (2014).
Stahelin, R. V. Could the Ebola virus matrix protein VP40 be a drug target? Exp. Opin. Therapeut. Targets 18, 115–120 (2014).
Gelinas, J. F., Clerzius, G., Shaw, E. & Gatignol, A. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 85, 8460–8466 (2011).
Schirmer, M. et al. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37 (2015).
Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files v.1.33 (Joshi, N. A. & Fass, J. N., 2011); https://github.com/najoshi/sickle
Nikolenko, S. I., Korobeynikov, A. I. & Alekseyev, M. A. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14(Suppl. 1), S7 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Acknowledgements
The authors thank Wu-Chun Cao, Hui Wang and Hong-Wu Yao for helpful discussions and C. Tomkins-Tinch for technical support. This work was supported by the China Ministry of Science and Technology (MOST) Project ‘973’ (grant numbers 2015CB910501 and 2015CB910503), the China National Grand S&T Special Project (grant numbers 2013ZX09304101 and 2015ZX09102024), the MOST Key Research and Development Program (2016YFC1200805), the Natural Science Foundation of China (grant numbers 31471253, 81590761 and U1435222), the National Hi-Tech R&D Program of China (‘863’ Program) (grant numbers 2014AA021501, 2014AA021505 and 2015AA020108), the Special Foundation of President for Ebola Virus Research from the Chinese Academy of Sciences (CAS) and the Yangfan Program from Beijing Municipal Administration of Hospitals (to H.Z.). W.S. was supported by the ‘Taishan Scholar’ project of Shandong Province. G.W. was supported by the President's International Fellowship Initiative from CAS. G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (grant no. 81321063).
Author information
Authors and Affiliations
Contributions
This study was conceived and designed by D.L., M.N., C.C., X.C.B. and G.F.G. Samples were collected and prepared by J.Q., H.J.L. and Z.P.X. Experiments were performed by Z.L., P.S.X. and H.X.X. The methods were developed and implemented by M.N., D.L. and C.C. The data were analysed by M.N., D.L., C.C., W.F.S., H.Y.W., H.X.X., Y.G.T., T.Y.G., Y.L., W.L. and J.W. The manuscript was prepared by D.L., M.N., C.C., W.F.S., G.W., X.C.B., H.Z. and G.F.G. This study was supervised by D.L., X.C.B., G.F.G., H.Z., J.M. and S.Q.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–9, Legends for Supplementary Tables 1–8 (PDF 3423 kb)
Supplementary Data
Supplementary Tables 1–8 (XLSX 251 kb)
Rights and permissions
About this article
Cite this article
Ni, M., Chen, C., Qian, J. et al. Intra-host dynamics of Ebola virus during 2014. Nat Microbiol 1, 16151 (2016). https://doi.org/10.1038/nmicrobiol.2016.151
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.151
This article is cited by
-
Convergence of two serotypes within the epidemic ST11 KPC-producing Klebsiella pneumoniae creates the “Perfect Storm” in a teaching hospital
BMC Genomics (2022)
-
Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients
Genome Medicine (2021)
-
Genomic surveillance of COVID-19 cases in Beijing
Nature Communications (2020)
-
Multiple approaches for massively parallel sequencing of SARS-CoV-2 genomes directly from clinical samples
Genome Medicine (2020)
-
Viral diseases meet omics: Time for systems virology
Science China Life Sciences (2018)