Abstract
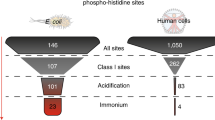
For decades, major difficulties in analyzing histidine phosphorylation have limited the study of phosphohistidine signaling. Here we report a method revealing widespread and abundant protein histidine phosphorylation in Escherichia coli. We generated an extensive E. coli phosphoproteome data set, in which a remarkably high percentage (∼10%) of phosphorylation sites are phosphohistidine sites. This resource should help enable a better understanding of the biological function of histidine phosphorylation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hunter, T. Cell 80, 225–236 (1995).
Laub, M.T. & Goulian, M. Annu. Rev. Genet. 41, 121–145 (2007).
Deutscher, J., Francke, C. & Postma, P.W. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006).
Fuhs, S.R. & Hunter, T. Curr. Opin. Cell Biol. 45, 8–16 (2017).
Boyer, P.D., Deluca, M., Ebner, K.E., Hultquist, D.E. & Peter, J.B. J. Biol. Chem. 237, PC3306–PC3308 (1962).
Kee, J.M., Oslund, R.C., Perlman, D.H. & Muir, T.W. Nat. Chem. Biol. 9, 416–421 (2013).
Fuhs, S.R. et al. Cell 162, 198–210 (2015).
Oslund, R.C. et al. J. Am. Chem. Soc. 136, 12899–12911 (2014).
Rush, J. et al. Nat. Biotechnol. 23, 94–101 (2005).
Sugiyama, N. et al. Mol. Cell. Proteomics 6, 1103–1109 (2007).
Kee, J.M. & Muir, T.W. ACS Chem. Biol. 7, 44–51 (2012).
Villén, J. & Gygi, S.P. Nat. Protoc. 3, 1630–1638 (2008).
Wei, Y.F. & Matthews, H.R. Methods Enzymol. 200, 388–414 (1991).
Ruprecht, B. et al. Mol. Cell. Proteomics 14, 205–215 (2015).
Mijakovic, I. & Macek, B. FEMS Microbiol. Rev. 36, 877–892 (2012).
Lin, M.H., Sugiyama, N. & Ishihama, Y. Sci. Signal. 8, rs10 (2015).
Zu, X.L., Besant, P.G., Imhof, A. & Attwood, P.V. Amino Acids 32, 347–357 (2007).
Medzihradszky, K.F., Phillipps, N.J., Senderowicz, L., Wang, P. & Turck, C.W. Protein Sci. 6, 1405–1411 (1997).
Capra, E.J. & Laub, M.T. Annu. Rev. Microbiol. 66, 325–347 (2012).
Li, G.W., Burkhardt, D., Gross, C. & Weissman, J.S. Cell 157, 624–635 (2014).
Ruprecht, B. et al. Methods Mol. Biol. 1550, 47–60 (2017).
Tyanova, S., Temu, T. & Cox, J. Nat. Protoc. 11, 2301–2319 (2016).
Cox, J. et al. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Tyanova, S. et al. Nat. Methods 13, 731–740 (2016).
Henderson, I.R., Owen, P. & Nataro, J.P. Mol. Microbiol. 33, 919–932 (1999).
Tusher, V.G., Tibshirani, R. & Chu, G. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Zolg, D.P. et al. Nat. Methods 14, 259–262 (2017).
Wei, Y.F. & Matthews, H.R. Methods Enzymol. 200, 388–414 (1991).
Acknowledgements
We thank R. Zheng and B. Kuster of the Technical University Munich, Freising, Germany, for providing us with synthetic peptides used in this study. This work was supported by the Roadmap Initiative Proteins@Work funded by the Netherlands Organization for Scientific Research (NWO) (to A.J.R.H.; project number 184.032.201) and by the MSMed program, funded by the European Union's Horizon 2020 Framework Programme (to A.J.R.H.; grant agreement number 686547). S.L. acknowledges support from the Netherlands Organization for Scientific Research (NWO) through a VIDI grant (project 723.013.008).
Author information
Authors and Affiliations
Contributions
C.M.P., M.-H.L., A.J.R.H. and S.L. designed the experiment. C.M.P. and M.-H.L. performed the experiments. C.M.P., M.-H.L. and S.L. performed data analysis. C.M.P., M.-H.L., A.J.R.H. and S.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Hydrolysis rates of synthetic pHis peptides
Ten peptides, corresponding to reported E. coli histidine tryptic phosphopeptides, were synthesized, chemically phosphorylated and subjected to acidic treatment, at pH 2.3 prior to LC-MS/MS analysis in triplicates. The label-free quantification function within the MaxQuant search algorithm was used to assess the hydrolysis rates (centre values representing the mean, error bars the standard deviation for n=3 samples). No significant loss of phosphohistidine was observed after 30 minutes of acid treatment, indicating that the method presented here, with a total time of 14 min for sample loading, washing and elution is suitable for the enrichment of histidine phosphorylation.
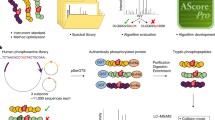
Supplementary Figure 2 Proof of principle of the experimental workflow
As a model system, synthetically generated phosphohistidine peptides were obtained from some standard proteins (Bovine Serum Albumin, Yeast Alcohol Dehydrogenase 1, Bovine catalase, Equine myoglobin, Human hemoglobin, Equine Cytochrome C and Bovine insulin) and enriched using a Fe3+-IMAC column. 500 μg of peptides were enriched and the retained phosphopeptides were analyzed by nanoLC-MS/MS. Data analysis using the MaxQuant search algorithm confirmed that the method presented here allows an efficient enrichment as well as preservation of histidine phosphopeptides (Table S1).
Supplementary Figure 3 Fe3+-IMAC UV chromatogram
Changes in the UV-trace at the outlet of the Fe3+-IMAC column attests of the efficient removal of impurities in our optimized sample preparation protocol. Indeed, in the case of the optimized protocol, the UV-abs signal quickly returns to baseline after non-retained peptides have been washed out. More importantly, the UV-abs signal and as such the amount of retained components by the Fe3+-IMAC resin decreases drastically. The same UV profiles were observed for the n=12 biological replicates. In terms of identifications, this translates into a ≈ 10 fold increase of the number of identified phosphopeptides per replicate. This drastic improvement can be explained by the fact that the presence of contaminants can severely impair the Fe3+-IMAC enrichment selectivity, as well as the quality of separation in liquid chromatography and/or ionization efficiency.
Supplementary Figure 4 Number of identified phosphopeptides
Total number of identified unique phosphopeptides (A) and class I phosphohistidine sites (B) as a function of the number of LC-MS/MS runs. Both are substantially higher than those reported in previously published studies, even after a single 90 minutes LC-MS/MS analysis (Figure S5).
Supplementary Figure 5 Comparison to previously reported data
Number of uniquely identified phosphopeptides, total Class I phosphosites, Class I pS/pT/pY and Class I pHis in our dataset compared with previously reported data. The method presented here shows a stunning increase in terms of identified histidine phosphorylation sites as well as all other phosphorylation events (on STY) within E. coli. The total number of class I phosphosites is around 10 times higher when compared to most published bacterial phosphoproteomics studies1-3, and 2 times higher when compared to the most recent study4, while using a significantly shorter analysis time.
1. Macek, B. et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics 7, 299-307, doi:10.1074/mcp.M700311-MCP200 (2008).
2. Qu, Y. et al. Automated immobilized metal affinity chromatography system for enrichment of Escherichia coli phosphoproteome. Electrophoresis 34, 1619-1626, doi:10.1002/elps.201200628 (2013).
3. Soares, N. C., Spat, P., Krug, K. & Macek, B. Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium. J Proteome Res 12, 2611-2621, doi:10.1021/pr3011843 (2013).
4. Lin, M. H., Sugiyama, N. & Ishihama, Y. Systematic profiling of the bacterial phosphoproteome reveals bacterium-specific features of phosphorylation. Sci Signal 8, rs10, doi:10.1126/scisignal.aaa3117 (2015).
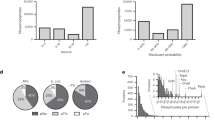
Supplementary Figure 6 Intensity distribution of detected endogenous E. coli class I S/T/Y/H phosphosites
Interestingly, and in contrast with higher organisms, no clear preference in the type of phosphorylation event is observed. Surprisingly, histidine phosphorylation even seems to be the most abundant phosphorylation event in E. coli, by a factor ≈ 2. The different intensities profiles derive from the totality of the n=12 biological replicates. This high abundancy attests of the good preservation of histidine phosphorylation through the enrichment and can be explained by the fact that histidine phosphorylation was mostly observed on several high abundant metabolic enzymes.
Supplementary Figure 7 Validation of pHis with synthetic peptides
Validation of histidine phosphorylation by comparison of the tandem MS spectrum derived from the endogenous peptide to spectra derived from synthetic peptides: example of the phosphosite H56 of the 30S ribosomal protein S10 (rpsJ). (A) MS2 spectrum acquired for the endogenous E. coli phosphopeptides. (B) MS2 spectrum acquired for the synthetic histidine phosphorylated peptide FTVLISPpHVNK. (C) MS2 spectrum acquired for the synthetic serine phosphorylated FTVLIpSPHVNK isoform. Comparison of the different spectra clearly confirms the correct assignment of histidine phosphorylation on the endogenous peptide (A). The 3 different MS2 profiles were each observed in 3 different spectra.
Supplementary Figure 8 Validation of pHis with synthetic peptides
Validation of histidine phosphorylation by comparison of the tandem MS spectrum derived from the endogenous peptide to spectra derived from synthetic peptides: example of the phosphosite H89 of the autonomous glycyl radical cofactor protein (grcA). (A) MS2 spectrum acquired for the endogenous E. coli phosphopeptide. (B) MS2 spectrum acquired for the synthetic histidine phosphorylated peptide RETLEDAVKpHPEK. (C) MS2 spectrum acquired for the synthetic threonine phosphorylated REpTLEDAVKHPEK isoform. Comparison of the different spectra clearly confirms the correct assignment of histidine phosphorylation of the endogenous peptide (A). The 3 different MS2 profiles were each observed in 3 different spectra.
Supplementary Figure 9 The phosphohistidine immonium ion can be used as a diagnostic ion
(A) Frequencies at which the ion of mass 190.0376 m/z was observed in the MS2 spectra of different type of samples (within 10 ppm mass tolerance). In proteome samples (E. coli digest), as well as in the boiled phosphoproteome samples, the 190.0376 m/z ion is barely present, while this pHis immonium ion was very frequently observed in the case of synthetic pHis peptides. This reveals that the 190.0376 m/z ion corresponds exclusively to the phosphohistidine immonium ion and as such can be used as a diagnostic tool. (B) The average intensity of endogenous E. coli phosphohistidine peptides for which the pHis immonium ion was observed is ≈ 2 times higher than the average intensity of all phosphohistidine containing peptides, showing that the occurrence of the pHis immonium ion is more prominent in high abundant phosphohistidine peptides. (C) The presence of the pHis immonium in endogenous E. coli phosphohistidine peptides’ MS2 spectra is highly dependent on the position of the phosphorylation within the peptide, with a clear bias toward the position 1. This is in parallel with the well-known phosphotyrosine immonium ion, for which a similar bias has been reported1.
1. Salek, M., Alonso, A., Pipkorn, R. & Lehmann, W. D. Analysis of protein tyrosine phosphorylation by nanoelectrospray ionization high-resolution tandem mass spectrometry and tyrosine-targeted product ion scanning. Anal Chem 75, 2724-2729 (2003).
Supplementary Figure 10 Phosphorylation motifs observed on endogenous E. coli phosphoproteins
While several S/T/Y phosphorylation motifs were identified, no histidine phosphorylation motif was statistically enriched (by the Motif-x software1, using the following parameters: 10 occurrences; 0.00005 significance and the E. coli proteome as background). This observation is in accordance with the fact that all known histidine kinases, phosphorylate distinct substrates2.
1. Schwartz, D. & Gygi, S. P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23, 1391-1398, doi:10.1038/nbt1146 (2005).
2. Capra, E. J. & Laub, M. T. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66, 325-347, doi:10.1146/annurev-micro-092611-150039 (2012).
Supplementary Figure 11 Assessment of the reproducibility of pHis peptide enrichment
After label-free quantification by the MaxQuant software, calculated Pearson correlation coefficients for the quantified class I phosphohistidine peptides (pink) in triplicate (each replicate corresponding to a different enrichment and different LC-MS/MS injection) were all above 0.93. This shows that our enrichment method is robust and enables reproducible quantification of histidine phosphorylation.
Supplementary Figure 12 Growth curve of glucose- or glycerol-fed E. coli
E. coli cells were collected at either mid-log phase or stationary phase according to OD600 absorbance.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Tables 4 and 6. (PDF 1290 kb)
Supplementary Table 1
List of identified phosphohistidine sites from chemically phosphorylated protein mixtures. (XLSX 18738 kb)
Supplementary Table 2
List of all identified phosphosites (STYH) from E. coli, including score, localization probability and precursor intensity in the different growth conditions. (XLSX 507 kb)
Supplementary Table 3
Comparison of identified phosphohistidine sites from antibody- and MS-based approaches. (XLSX 30 kb)
Supplementary Table 5
List of quantified phosphosites (STYH) and statistical analysis of detected changes between exponential and stationary phases of glycerol-fed E. coli. (XLSX 162 kb)
Source data
Rights and permissions
About this article
Cite this article
Potel, C., Lin, MH., Heck, A. et al. Widespread bacterial protein histidine phosphorylation revealed by mass spectrometry-based proteomics. Nat Methods 15, 187–190 (2018). https://doi.org/10.1038/nmeth.4580
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4580
This article is cited by
-
Control of protein stability by post-translational modifications
Nature Communications (2023)
-
pHisPred: a tool for the identification of histidine phosphorylation sites by integrating amino acid patterns and properties
BMC Bioinformatics (2022)
-
Histidine phosphorylation in human cells; a needle or phantom in the haystack?
Nature Methods (2022)
-
Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome
Nature Methods (2022)
-
Construction and analysis of an integrated biological network of Escherichia coli
Systems Microbiology and Biomanufacturing (2022)