Abstract
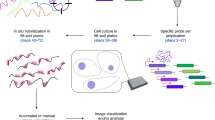
We report a high-throughput screening method that allows diverse genotypes and corresponding phenotypes to be imaged in individual cells. We achieve genotyping by introducing barcoded genetic variants into cells as pooled libraries and reading the barcodes out using massively multiplexed fluorescence in situ hybridization. To demonstrate the power of image-based pooled screening, we identified brighter and more photostable variants of the fluorescent protein YFAST among 60,000 variants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Cadwell, R.C. & Joyce, G.F. PCR Methods Appl. 2, 28–33 (1992).
Kosuri, S. & Church, G.M. Nat. Methods 11, 499–507 (2014).
Zhang, J., Campbell, R.E., Ting, A.Y. & Tsien, R.Y. Nat. Rev. Mol. Cell Biol. 3, 906–918 (2002).
Grotjohann, T. et al. Nature 478, 204–208 (2011).
Brakemann, T. et al. Nat. Biotechnol. 29, 942–947 (2011).
Shaner, N.C. et al. Nat. Methods 5, 545–551 (2008).
Davis, L.M., Lubbeck, J.L., Dean, K.M., Palmer, A.E. & Jimenez, R. Lab Chip 13, 2320–2327 (2013).
Dean, K.M. et al. Integr. Biol. 7, 263–273 (2015).
Shalem, O., Sanjana, N.E. & Zhang, F. Nat. Rev. Genet. 16, 299–311 (2015).
Dixit, A. et al. Cell 167, 1853–1866.e17 (2016).
Adamson, B. et al. Cell 167, 1867–1882.e21 (2016).
Jaitin, D.A. et al. Cell 167, 1883–1896.e15 (2016).
Datlinger, P. et al. Nat. Methods 14, 297–301 (2017).
Plamont, M.-A. Proc. Natl. Acad. Sci. USA 113, 497–502 (2016).
Chen, K.H., Boettiger, A.N., Moffitt, J.R., Wang, S. & Zhuang, X. Science 348, aaa6090 (2015).
Moffitt, J.R. et al. Proc. Natl. Acad. Sci. USA 113, 11046–11051 (2016).
Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Science 280, 585–590 (1998).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Nat. Methods 5, 877–879 (2008).
Subach, O.M., Cranfill, P.J., Davidson, M.W. & Verkhusha, V.V. PLoS One 6, e28674 (2011).
Wang, S., Moffitt, J.R., Dempsey, G.T., Xie, X.S. & Zhuang, X. Proc. Natl. Acad. Sci. USA 111, 8452–8457 (2014).
Zhang, Z., Revyakin, A., Grimm, J.B., Lavis, L.D. & Tjian, R. eLife 3, e01775 (2014).
Gibson, D.G. et al. Nat. Methods 6, 343–345 (2009).
Lutz, R. & Bujard, H. Nucleic Acids Res. 25, 1203–1210 (1997).
Kivioja, T. et al. Nat. Methods 9, 72–74 (2011).
Shiroguchi, K., Jia, T.Z., Sims, P.A. & Xie, X.S. Proc. Natl. Acad. Sci. USA 109, 1347–1352 (2012).
Getzoff, E.D., Gutwin, K.N. & Genick, U.K. Nat. Struct. Biol. 10, 663–668 (2003).
Shaffer, S.M., Wu, M.-T., Levesque, M.J. & Raj, A. PLoS One 8, e75120 (2013).
Rasnik, I., McKinney, S.A. & Ha, T. Nat. Methods 3, 891–893 (2006).
Emanuel, G., Moffitt, J.R. & Zhuang, X. Protocol Exchange https://doi.org/10.1038/protex.2017.109 (2017).
Acknowledgements
We thank H. Babcock for instrumentation advice and many readers for participating in the discussion of the preprint of this paper that we deposited on bioRxiv. This work was supported in part by the NIH. X.Z. is an HHMI investigator.
Author information
Authors and Affiliations
Contributions
G.E., J.R.M., and X.Z. conceived the study and designed the experiments. G.E. performed experiments and analyzed data. G.E., J.R.M., and X.Z. interpreted the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.E., J.R.M., and X.Z. are inventors on a patent applied for by Harvard University that covers the screening method described here.
Integrated supplementary information
Supplementary Figure 1 Distributions of the ratio of readout 0 intensity to the readout 1 intensity.
Histogram of the natural logarithm of the ratio of readout 0 intensity to readout 1 intensity for bit 1 for only those cells with assigned barcodes that match the valid barcodes that are determined to be in the library by sequencing. The histogram was fit to a sum of two skewed Gaussians and each fit Gaussian is depicted (green and magenta dashed lines). The overlap between the two Gaussian curves corresponds to 0.07% of the total number of cells presented here. The dotted cyan line depicts the bit-calling threshold. Cells above the bit-calling threshold were assigned a bit value of “0” for this bit while cells below the bit-calling threshold were assigned a bit value of “1”.
Supplementary Figure 2 Reversible and biphasic photobleaching kinetics of YFAST.
The normalized fluorescence intensity (green crosses) upon intermittent illumination with 488-nm light and a fit to a double exponential decay (purple line). E. coli expressing BFP-YFAST were adhered to a glass coverslip, immersed in 10 μM HMBR in PBS and imaged at 4-ms time resolution. The YFAST fluorescence intensity for each cell is normalized by the BFP fluorescence and averaged over multiple cells in the imaged area. The 488-nm illumination was switched on and off with a period of 2 seconds. Intensity values of zero represent the period of time when the illumination was off.
Supplementary Figure 3 Additional quantifications of the YFAST library screen and comparisons between library measurements and isolated mutant measurements.
(a) Scatter plot of the apparent fractional amplitudes of the fast bleaching component and the amplitudes of the slow bleaching component for different YFAST mutants in all of the libraries imaged. Each point depicts the median values of all cells associated with one mutant. The amplitudes of the slow bleaching component are normalized to that of the original YFAST. The apparent fractional amplitude of the fast bleaching component is defined as the amplitude of the fast bleaching component divided by the sum of the amplitudes of the fast and slow bleaching components. Here only the mutants that contain at least 10 imaged cells are depicted. The library measurements of the original YFAST and three selected mutants are indicated by the green star and colored circles, respectively. The color scheme is as in Fig. 3. (b) Scatter plot of the apparent fractional amplitudes of the fast bleaching component for each mutant measured in two replicate library measurements containing a subset of the 60,000 mutants. Each point represents the median of all cells corresponding to each mutant and is colored by the minimum number of cells measured in either replicate. Only the mutants with a slow component at least half as bright as the slow component for the original YFAST and that contain at least 10 imaged cells in each replicate are depicted. (c) The apparent fractional amplitude of the fast bleaching component for the original YFAST and the selected mutants determined from the isolated mutant measurements at the same time resolution as the library measurements shown together with the results obtained from the library measurements. Solid circles represent the median from the library measurements (N=420, 136, 32, 220 cells for the original YFAST, mutant 1, mutant 2, and mutant 3, respectively). Diamonds represent the mean from the isolated mutant measurements (N = 12, 5, 3, and 3 technical replicates for original YFAST, mutants 1, mutant 2, and mutant 3, respectively). Error bars represent SEM and individual measurements are depicted as gray crosses when N is less than 10. (d) The fluorescence intensity as a function of illumination time for the original YFAST and the three selected mutants (color-coded as in (a)) measured in isolation at the 4-ms time resolution. The fluorescence intensity is averaged over multiple cells. (e and f) The fractional amplitude (e) and photobleaching rate (f) of the fast bleaching component for the original YFAST and the selected mutants determined from the isolated mutant measurements shown in (d). Diamonds depict the mean and error bars represent SEM. Individual measurements are depicted as gray crosses (N = 3 replicates for the original YFAST and the three mutants). The amplitudes and rate constants of both fast and slow bleaching components are determined from these fast-time resolution measurements by a double exponential fit. The fractional amplitudes and the rate constants of the fast bleaching component are shown in the plot here, and the amplitudes and rate constants of the slow bleaching component are shown in Fig. 3f, g.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–2 and Supplementary Note 1 (PDF 878 kb)
Supplementary Protocol
Library construction and imaging for high-throughput image-based screening using multiplexed FISH. (PDF 376 kb)
Supplementary Software
Software for analysis of image data and sequencing data. (ZIP 129 kb)
Rights and permissions
About this article
Cite this article
Emanuel, G., Moffitt, J. & Zhuang, X. High-throughput, image-based screening of pooled genetic-variant libraries. Nat Methods 14, 1159–1162 (2017). https://doi.org/10.1038/nmeth.4495
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4495
This article is cited by
-
Emerging tools for uncovering genetic and transcriptomic heterogeneities in bacteria
Biophysical Reviews (2024)
-
Spatial transcriptomics for profiling the tropism of viral vectors in tissues
Nature Biotechnology (2023)
-
A synthetic population-level oscillator in non-microfluidic environments
Communications Biology (2023)
-
Synthetic Micrographs of Bacteria (SyMBac) allows accurate segmentation of bacterial cells using deep neural networks
BMC Biology (2022)
-
The emerging landscape of spatial profiling technologies
Nature Reviews Genetics (2022)