Abstract
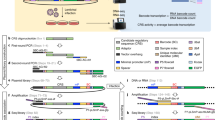
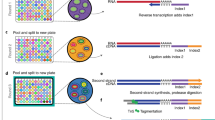
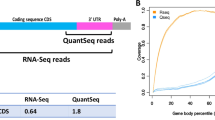
We describe a method called modular, early-tagged amplification (META) RNA profiling that can quantify a broad panel of microRNAs or mRNAs simultaneously across many samples and requires far less sequence depth than existing digital profiling technologies. The method assigns quantitative tags during reverse transcription to permit up-front sample pooling before competitive amplification and deep sequencing. This simple, scalable and inexpensive approach improves the practicality of large-scale gene expression studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Nagalakshmi, U. et al. Science 320, 1344–1349 (2008).
Wilhelm, B.T. et al. Nature 453, 1239–1243 (2008).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Nat. Methods 5, 621–628 (2008).
Cloonan, N. et al. Nat. Methods 5, 613–619 (2008).
Wang, Z., Gerstein, M. & Snyder, M. Nat. Rev. Genet. 10, 57–63 (2009).
Velculescu, V.E., Zhang, L., Vogelstein, B. & Kinzler, K.W. Science 270, 484–487 (1995).
Schena, M., Shalon, D., Davis, R.W. & Brown, P.O. Science 270, 467–470 (1995).
Griffiths-Jones, S., Grocock, R.J., van Dongen, S., Bateman, A. & Enright, A.J. Nucleic Acids Res. 34, D140–D144 (2006).
Gaur, A. et al. Cancer Res. 67, 2456–2468 (2007).
Mestdagh, P. et al. Nucleic Acids Res. 39, e136 (2011).
Liang, Y., Ridzon, D., Wong, L. & Chen, C. BMC Genomics 8, 166 (2007).
Willems, E., Leyns, L. & Vandesompele, J. Anal. Biochem. 379, 127–129 (2008).
Mestdagh, P. et al. Genome Biol. 10, R64 (2009).
D'haene, B., Mestdagh, P., Hellemans, J. & Vandesompele, J. Methods Mol. Biol. 822, 261–272 (2012).
Blower, P.E. et al. Mol. Cancer Ther. 6, 1483–1491 (2007).
Liu, H. et al. Mol. Cancer Ther. 9, 1080–1091 (2010).
Søkilde, R. et al. Mol. Cancer Ther. 10, 375–384 (2011).
Canales, R.D. et al. Nat. Biotechnol. 24, 1115–1122 (2006).
Shippy, R. et al. Nat. Biotechnol. 24, 1123–1131 (2006).
Paul, S. & Amundson, S.A. Int. J. Radiat. Oncol. Biol. Phys. 71, 1236–1244 (2008).
Paul, S. et al. Radiat. Res. 175, 257–265 (2011).
Chen, C. et al. Nucleic Acids Res. 33, e179 (2005).
Acknowledgements
We thank J. Deluca and A. Blanchard for help with oligonucleotide synthesis; S. Mane and N. DaSilva for assistance with next-generation sequencing; P. Gareiss and M. Salcius for help with robotic liquid handling; S. Holbeck (US National Cancer Institute) for providing RNA from the NCI-60 cancer cell lines; M. Saba for assistance with creating figures; and J. Steitz, D. Brash and P. Glazer for valuable comments on the manuscript. Research was supported by the Yale Cancer Center, The Honorable Tina Brozman Foundation, a Rudolph Anderson fellowship, a Leslie Warner fellowship, and Clinical and Translational Science Award grants UL1 TR000142 and KL2 TR000140 from the National Center for Advancing Translational Sciences, a component of the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
All authors contributed to planning of experiments, analysis of data and writing of the paper. A.A.P. conceived of the design and supervised the study. A.N. performed the majority of experimental work, and A.B. performed experiments related to radiation biodosimetry.
Corresponding author
Ethics declarations
Competing interests
A provisional patent application has been filed covering the described method, with A.A.P. listed as an inventor.
Integrated supplementary information
Supplementary Figure 1 Accuracy of multiplexed RNA quantitation.
(a) A heat map displays a 9,216 pixel image of a rose based on measurements of 96 synthetic miRNAs and control RNAs mixed in specified proportions within 96 samples. Mean values of 2 experimental replicates are shown, each sequenced using a high-capacity 318 chip. Normalization is described in Online Methods. RNAs are in the same order as listed in Supplementary Table 2. (b) A similar heat map generated from 2 replicates using lower-capacity 314 chips. With fewer sequence counts, the image appears more grainy (like a photograph taken in low light) but still exhibits a strong quantitative signal above noise (c) Concordance between the amount of synthetic RNA added to a sample and its measured level using 318 or (d) 314 chips. Fold-change is relative to the mean for each RNA. (e) Effect of sequence depth on quantitative accuracy, defined by the Pearson correlation coefficient between known and measured RNA levels.
Supplementary Figure 2 Creation of the synthetic RNA heat map image.
A digital photograph of a rose (taken by AAP) was reduced to 96 x 96 pixel resolution. Two new images were then created by segregating the red from non-red regions using the color-range selection function in Adobe Photoshop. These two images were converted to 8-bit greyscale bitmap files, from which the numerical grey level for each pixel could be extracted (0 = black, 255 = white). After converting the values from the non-red area to negative numbers, all values (from –255 to 255) were transformed using the following function: f(x) = 5*2^(x/76.763). Resulting values ranged from 0.5 to 50, and were used to specify the number of microliters of a particular synthetic RNA (pixel row) to be dispensed by the robot into a given sample tube (pixel column). Extra buffer was added to equalize the final volume in all 96 tubes. Shown is a theoretical heat map that would be generated if RNAs were perfectly dispensed and measured. An input error was discovered only after the RNAs had been measured: the robot had not dispensed any volumes less than 1 μL, and instead for all such input values, had shifted the decimal point one place to the right.
Supplementary Figure 3 Repeatability of synthetic RNA measurements.
(a) To supplement the two replicate 318 chip measurements presented in Supplementary Figure 1, a third technical replicate was performed (3.36 million filtered reads). Primer mixes with different tags were used to reverse-transcribe the same sample in different replicates. Coefficients of variation (CVs) were calculated between the 3 replicate measurements for each data bin after normalization as described in Online Methods, but without log-transformation. Distributions of these CV values are presented as box and whiskers plots, grouped by RNA. The median (horizontal line), interquartile range (box), and 10th and 90th percentile values (whiskers) are indicated. RNAs are listed in the same order as in Supplementary Table 2. (b) The same CV distributions are plotted after filtering to remove values that had a raw read count of 10 or less in any of the 3 replicates.
Supplementary Figure 4 Complete heat map of miRNA measurements from normal human tissues compared to qRT-PCR data.
A heat map with divided pixels compares levels of 90 miRNAs measured as 3 technical replicates from 20 normal human tissues to published qRT-PCR measurements10. Both data sets were standardized as previously described12-14. This Figure shows the full data set that was partially displayed in Figure 2a due to space constraints.
Supplementary Figure 5 miRNA levels measured from NCI-60 cancer cell lines compared to qRT-PCR data.
(a) A heat map with divided pixels compares levels of 54 miRNAs measured in a single experiment from 58 cell lines to published qRT-PCR measurements9. Consistent with prior studies9, 15-17, we found several miRNAs that showed poor expression in the majority of NCI-60 lines. Such miRNAs were excluded from consideration if in more than 85% of cell lines, they had published Cq values > 33 or our measurements produced raw read counts < 10. Two cell lines were omitted: MDA-MB-468 because it was not included in published data, and HCC-2998 because its RNA was degraded. (b) Heat map of correlation coefficients of miRNA levels measured by META RNA profiling vs. qRT-PCR from the same cell line (diagonal) or between different cell lines (off-diagonal). Cell lines are grouped by tissue of origin in order to highlight off-diagonal correlations among lines derived from the same cancer type.
Supplementary Figure 6 NCI-60 miRNA levels compared to pin-spotted microarray data.
Data format is identical to Supplementary Fig. 5. (a) Heat map with divided pixels compares miRNA levels measured from NCI-60 cell lines to the custom pin-spotted microarray data of Blower et al.15. (b) Heat map of correlation coefficients of miRNA levels measured by META RNA profiling vs. pin-spotted microarray from the same cell line (diagonal) or between different cell lines (off-diagonal).
Supplementary Figure 7 NCI-60 miRNA levels compared to Agilent microarray data.
Data format is identical to Supplementary Fig. 5. (a) Heat map with divided pixels compares miRNA levels measured from NCI-60 cell lines to Agilent Human microRNA Microarray V2 data of Liu et al.16. (b) Heat map of correlation coefficients of miRNA levels measured by META RNA profiling vs. Agilent microarray from the same cell line (diagonal) or between different cell lines (off-diagonal).
Supplementary Figure 8 NCI-60 miRNA levels compared to LNA microarray data.
Data format is identical to Supplementary Fig. 5. (a) Heat map with divided pixels compares miRNA levels measured from NCI-60 cell lines to Exiqon LNA-enhanced miRCURY Dx microarray data of Sokilde et al.17. (b) Heat map of correlation coefficients of miRNA levels measured by META RNA profiling vs. LNA microarray from the same cell line (diagonal) or between different cell lines (off-diagonal).
Supplementary Figure 9 Comparison of several miRNA-profiling platforms.
(a) miRNA levels were measured in total RNA derived from normal human brain and liver using five orthogonal platforms, including META RNA profiling. Data from the other four platforms were reported by independent laboratories. Values for log2(fold-difference) between miRNA levels in brain vs. liver as measured by different platforms are displayed in the heat map. Analysis was restricted to miRNAs that were common to all assay panels. An miRNA was excluded if its level was below the limit of detection in both samples for a given platform. META RNA profiling values are means of 3 technical replicates. (b) Pairwise correlation of fold-difference of miRNA levels between brain and liver as measured by META RNA profiling vs. four orthogonal platforms. (c) Pairwise R2 values for all platform combinations. External data sets were downloaded from mirnabodymap.org for Taqman qRT-PCR10 and from Gene Expression Omnibus for Illumina RNA-Seq (GSE49816), for Affymetrix array (GSE49661) and for NanoString (GSE49600).
Supplementary Figure 10 Absolute quantitation of miRNAs in human tissues.
By normalizing relative to a co-amplified quantitative reference standard sample containing ~15,000 synthetic copies of each miRNA species, the absolute miRNA concentration could be estimated. Total RNA input was 50 ng per tissue sample. Values were derived from the mean of 3 replicate RT reactions, which were pooled for singleplex PCR. Hsa-mir-381 was excluded from the analysis because it amplified poorly. The shade scale indicates miRNA abundance, and an embedded histogram indicates the frequency distribution of these values on the same scale.
Supplementary Figure 11 Radiation-induced gene expression in blood samples from 18 individuals.
Changes in expression levels of a panel of radiation-responsive genes were measured 24 hours after ex vivo exposure of blood samples to ionizing radiation doses of 0 to 8 Gy. All 108 samples from 18 subjects were processed and measured in parallel in two replicate experiments. Heat maps show standardized gene expression values (row Z-score). Mean centering and autoscaling were performed separately across samples from each subject.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–10, Supplementary Notes 1–4 and Supplementary Protocol (PDF 2221 kb)
Source data
Rights and permissions
About this article
Cite this article
Narayan, A., Bommakanti, A. & Patel, A. High-throughput RNA profiling via up-front sample parallelization. Nat Methods 12, 343–346 (2015). https://doi.org/10.1038/nmeth.3311
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3311
This article is cited by
-
MicroRNAs and Regeneration in Animal Models of CNS Disorders
Neurochemical Research (2020)
-
Deep sequencing and miRNA profiles in alcohol-induced neuroinflammation and the TLR4 response in mice cerebral cortex
Scientific Reports (2018)
-
Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification
Nature Biomedical Engineering (2017)
-
Multiplex enrichment quantitative PCR (ME-qPCR): a high-throughput, highly sensitive detection method for GMO identification
Analytical and Bioanalytical Chemistry (2017)
-
An integrated miRNA functional screening and target validation method for organ morphogenesis
Scientific Reports (2016)