Abstract
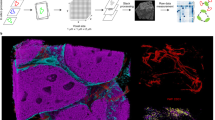
Mass cytometry enables high-dimensional, single-cell analysis of cell type and state. In mass cytometry, rare earth metals are used as reporters on antibodies. Analysis of metal abundances using the mass cytometer allows determination of marker expression in individual cells. Mass cytometry has previously been applied only to cell suspensions. To gain spatial information, we have coupled immunohistochemical and immunocytochemical methods with high-resolution laser ablation to CyTOF mass cytometry. This approach enables the simultaneous imaging of 32 proteins and protein modifications at subcellular resolution; with the availability of additional isotopes, measurement of over 100 markers will be possible. We applied imaging mass cytometry to human breast cancer samples, allowing delineation of cell subpopulations and cell-cell interactions and highlighting tumor heterogeneity. Imaging mass cytometry complements existing imaging approaches. It will enable basic studies of tissue heterogeneity and function and support the transition of medicine toward individualized molecularly targeted diagnosis and therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kherlopian, A.R. et al. A review of imaging techniques for systems biology. BMC Syst. Biol. 2, 74 (2008).
Lichtman, J.W. & Conchello, J.A. Fluorescence microscopy. Nat. Methods 2, 910–919 (2005).
Lubeck, E. & Cai, L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 9, 743–748 (2012).
Giepmans, B.N., Adams, S.R., Ellisman, M.H. & Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006).
Langer-Safer, P.R., Levine, M. & Ward, D.C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 79, 4381–4385 (1982).
Robertson, D., Savage, K., Reis-Filho, J.S. & Isacke, C.M. Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol. 9, 13 (2008).
Tsurui, H. et al. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J. Histochem. Cytochem. 48, 653–662 (2000).
Gerdes, M.J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 110, 11982–11987 (2013).
Schubert, W. et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 24, 1270–1278 (2006).
Wählby, C., Erlandsson, F., Bengtsson, E. & Zetterberg, A. Sequential immunofluorescence staining and image analysis for detection of large numbers of antigens in individual cell nuclei. Cytometry 47, 32–41 (2002).
Martell, J.D. et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30, 1143–1148 (2012).
Giepmans, B.N., Deerinck, T.J., Smarr, B.L., Jones, Y.Z. & Ellisman, M.H. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods 2, 743–749 (2005).
Cornett, D.S., Reyzer, M.L., Chaurand, P. & Caprioli, R.M. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods 4, 828–833 (2007).
Schober, Y., Guenther, S., Spengler, B. & Römpp, A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 84, 6293–6297 (2012).
McDonnell, L.A. & Heeren, R.M. Imaging mass spectrometry. Mass Spectrom. Rev. 26, 606–643 (2007).
Thiery, G. et al. Multiplex target protein imaging in tissue sections by mass spectrometry—TAMSIM. Rapid Commun. Mass Spectrom. 21, 823–829 (2007).
Qin, Z., Caruso, J.A., Lai, B., Matusch, A. & Becker, J.S. Trace metal imaging with high spatial resolution: applications in biomedicine. Metallomics 3, 28–37 (2011).
Zhang, D.S. et al. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481, 520–524 (2012).
Becker, J.S. et al. Bioimaging of metals and biomolecules in mouse heart by laser ablation inductively coupled plasma mass spectrometry and secondary ion mass spectrometry. Anal. Chem. 82, 9528–9533 (2010).
Koch, J. & Günther, D. Review of the state-of-the-art of laser ablation inductively coupled plasma mass spectrometry. Appl. Spectrosc. 65, 155–162 (2011).
Seuma, J. et al. Combination of immunohistochemistry and laser ablation ICP mass spectrometry for imaging of cancer biomarkers. Proteomics 8, 3775–3784 (2008).
Giesen, C. et al. Multiplexed immunohistochemical detection of tumor markers in breast cancer tissue using laser ablation inductively coupled plasma mass spectrometry. Anal. Chem. 83, 8177–8183 (2011).
Bandura, D.R. et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813–6822 (2009).
Lou, X. et al. Polymer-based elemental tags for sensitive bioassays. Angew. Chem. Int. Ed. Engl. 46, 6111–6114 (2007).
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012).
Hanahan, D. & Coussens, L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Psaila, B. & Lyden, D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293 (2009).
Hanahan, D. & Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Wang, H.A.O. et al. Fast chemical imaging at high spatial resolution by laser ablation inductively coupled plasma mass spectrometry. Anal. Chem. 85, 10107–10116 (2013).
Elenbaas, B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15, 50–65 (2001).
Perou, C.M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Sims, A.H., Howell, A., Howell, S.J. & Clarke, R.B. Origins of breast cancer subtypes and therapeutic implications. Nat. Clin. Pract. Oncol. 4, 516–525 (2007).
Nowell, P.C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Lock, F.E. et al. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 32, 5210–5219 (2013).
Davila, E. & Amazon, K. The clinical importance of the heterogeneity of HER2 neu. Case Rep. Oncol. 3, 268–271 (2010).
Theurillat, J.P. et al. NY-BR-1 protein expression in breast carcinoma: a mammary gland differentiation antigen as target for cancer immunotherapy. Cancer Immunol. Immunother. 56, 1723–1731 (2007).
Blurry, R.W. Immunocytochemistry, a Practical Guide for Biomedical Research (Springer, 2010).
Currie, L.A. Nomenclature in evaluation of analytical methods including detection and quantification capabilities (IUPAC recommendations 1995). Pure Appl. Chem. 67, 1699–1723 (1995).
Meyer, F. Topographic distance and watershed lines. Signal Processing 38, 113–125 (1994).
Shapiro, L.G. & Stockman, G.C. Computer Vision (Prentice Hall, 2001).
Kamentsky, L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011).
Acknowledgements
We thank M. Storz for preparing the histological slides and the TMA sections; S. Dettwiler, A. Bohnert, A. Fitsche; the entire Trace Element and Micro Analysis group at ETH Zürich for their experimental support and discussions; N. Daga and C. von Mering for their feedback on data analysis; the ETHZ LAC workshop for their support in design and construction of the laser ablation chamber; and the Lehner and Luschnig groups for giving us access to their immunofluorescence microscopes. This work was supported by a Society in Science, The Branco Weiss Fellowship, administered by the ETH Zürich (C.G.); the Swiss National Science Foundation (SNSF) project grants 200021-119779 (H.A.O.W.), 200021-119779 (D. Günther), 31003A-143877 (D. Günther) and 31003A-143877 (B.B.); an ETH Zürich Pioneer Fellowship (H.A.O.W.); the SystemsX PhosphoNet-PPM grant (P.J.W. and B.B.); a Baugarten Foundation grant (SGGP) (P.J.W.); a EU VIGOR++ project FP7/2007-2013, #270379 (P.J.S. and J.M.B.); an SNSF R'Equip grant 316030-139220 (B.B.); an SNSF Assistant Professorship grant PP00P3-144874 (B.B.); a Swiss Cancer League grant (B.B.); and funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement no. 336921 (B.B.).
Author information
Authors and Affiliations
Contributions
C.G., H.A.O.W., N.Z., A.J., D. Grolimund, Z.V., P.J.W., D. Günther and B.B. designed and performed experiments. H.A.O.W., D. Grolimund and B.H. established the tissue laser ablation system. C.G., H.A.O.W., D.S., N.Z., A.J., P.J.S. and D. Grolimund performed data analysis. S.B., Z.V. and P.J.W. assembled, provided and classified tumor samples. D.S., P.J.S. and J.M.B. arranged image analysis and single-cell segmentation. C.G., H.A.O.W., D.S., N.Z., P.J.W., D. Grolimund, D. Günther and B.B. prepared the figures and wrote the manuscript. D. Günther and B.B. conceived of and supervised the project. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Fluidigm (formerly DVS Sciences) has an option to license imaging mass cytometry technology from D. Günther represented by ETH Zürich, which includes a related funded research collaboration benefiting the D. Günther and B.B. labs.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1–6 and Supplementary Notes 1–3 (PDF 4881 kb)
Supplementary Data
Spade trees generated for the analysed breast cancer images (ZIP 14536 kb)
Supplementary Software
Imaging mass cytometry software (ZIP 100 kb)
Rights and permissions
About this article
Cite this article
Giesen, C., Wang, H., Schapiro, D. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422 (2014). https://doi.org/10.1038/nmeth.2869
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2869
This article is cited by
-
MAPS: pathologist-level cell type annotation from tissue images through machine learning
Nature Communications (2024)
-
Charting multicellular tissue structure cell-to-cell
Nature Genetics (2024)
-
Microenvironmental reorganization in brain tumors following radiotherapy and recurrence revealed by hyperplexed immunofluorescence imaging
Nature Communications (2024)
-
Mapping the single cell spatial immune landscapes of the melanoma microenvironment
Clinical & Experimental Metastasis (2024)
-
Imaging mass cytometry analysis of Becker muscular dystrophy muscle samples reveals different stages of muscle degeneration
Scientific Reports (2024)