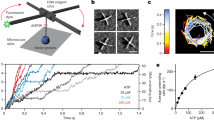
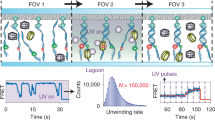
Abstract
Single-molecule measurements of DNA twist and extension have been used to reveal physical properties of the double helix and to characterize structural dynamics and mechanochemistry in nucleoprotein complexes. However, the spatiotemporal resolution of twist measurements has been limited by the use of angular probes with high rotational drag, which prevents detection of short-lived intermediates or small angular steps. We introduce gold rotor bead tracking (AuRBT), which yields >100× improvement in time resolution over previous techniques. AuRBT employs gold nanoparticles as bright low-drag rotational and extensional probes, which are monitored by instrumentation that combines magnetic tweezers with objective-side evanescent darkfield microscopy. Our analysis of high-speed structural dynamics of DNA gyrase using AuRBT revealed an unanticipated transient intermediate. AuRBT also enables direct measurements of DNA torque with >50× shorter integration times than previous techniques; we demonstrated high-resolution torque spectroscopy by mapping the conformational landscape of a Z-forming DNA sequence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Moffitt, J.R., Chemla, Y.R., Smith, S.B. & Bustamante, C. Recent advances in optical tweezers. Annu. Rev. Biochem. 77, 205–228 (2008).
Neuman, K.C. & Nagy, A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008).
Bryant, Z., Oberstrass, F.C. & Basu, A. Recent developments in single-molecule DNA mechanics. Curr. Opin. Struct. Biol. 22, 304–312 (2012).
Vologodskii, A. Determining protein-induced DNA bending in force-extension experiments: theoretical analysis. Biophys. J. 96, 3591–3599 (2009).
Nollmann, M., Crisona, N.J. & Arimondo, P.B. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie 89, 490–499 (2007).
Basu, A., Schoeffler, A.J., Berger, J.M. & Bryant, Z. ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat. Struct. Mol. Biol. 19, 538–546 (2012).
Gore, J. et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature 439, 100–104 (2006).
Killian, J.L., Li, M., Sheinin, M.Y. & Wang, M.D. Recent advances in single molecule studies of nucleosomes. Curr. Opin. Struct. Biol. 22, 80–87 (2012).
Revyakin, A., Liu, C., Ebright, R.H. & Strick, T.R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006).
Lee, M., Lipfert, J., Sanchez, H., Wyman, C. & Dekker, N.H. Structural and torsional properties of the RAD51-dsDNA nucleoprotein filament. Nucleic Acids Res. 41, 7023–7030 (2013).
Arata, H. et al. Direct observation of twisting steps during Rad51 polymerization on DNA. Proc. Natl. Acad. Sci. USA 106, 19239–19244 (2009).
Lipfert, J., Wiggin, M., Kerssemakers, J.W., Pedaci, F. & Dekker, N.H. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat. Commun. 2, 439 (2011).
Schoeffler, A.J. & Berger, J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 (2008).
Harada, Y. et al. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature 409, 113–115 (2001).
Abbondanzieri, E.A., Greenleaf, W.J., Shaevitz, J.W., Landick, R. & Block, S.M. Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005).
Li, G., Levitus, M., Bustamante, C. & Widom, J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 (2005).
Mosconi, F., Allemand, J.F. & Croquette, V. Soft magnetic tweezers: a proof of principle. Rev. Sci. Instrum. 82, 034302 (2011).
Forth, S., Sheinin, M.Y., Inman, J. & Wang, M.D. Torque measurement at the single-molecule level. Annual Review of Biophysics 42, 583–604 (2013).
Bryant, Z. et al. Structural transitions and elasticity from torque measurements on DNA. Nature 424, 338–341 (2003).
Janssen, X.J. et al. Electromagnetic torque tweezers: a versatile approach for measurement of single-molecule twist and torque. Nano Lett. 12, 3634–3639 (2012).
Lipfert, J., Kerssemakers, J.W., Jager, T. & Dekker, N.H. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods 7, 977–980 (2010).
Oberstrass, F.C., Fernandes, L.E. & Bryant, Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc. Natl. Acad. Sci. USA 109, 6106–6111 (2012).
Forth, S. et al. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 100, 148301 (2008).
Celedon, A. et al. Magnetic tweezers measurement of single molecule torque. Nano Lett. 9, 1720–1725 (2009).
Oberstrass, F.C., Fernandes, L.E., Lebel, P. & Bryant, Z. Torque Spectroscopy of DNA: Base-Pair Stability, Boundary Effects, Backbending, and Breathing Dynamics. Phys. Rev. Lett. 110, 178103 (2013).
Patel, S.S. & Donmez, I. Mechanisms of helicases. J. Biol. Chem. 281, 18265–18268 (2006).
Deufel, C., Forth, S., Simmons, C.R., Dejgosha, S. & Wang, M.D. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat. Methods 4, 223–225 (2007).
Zocchi, G. Proteins unfold in steps. Proc. Natl. Acad. Sci. USA 94, 10647–10651 (1997).
Liu, R., Garcia-Manyes, S., Sarkar, A., Badilla, C.L. & Fernandez, J.M. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys. J. 96, 3810–3821 (2009).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. Jr. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
Dunn, A.R. & Spudich, J.A. Dynamics of the unbound head during myosin V processive translocation. Nat. Struct. Mol. Biol. 14, 246–248 (2007).
Lindner, M. et al. Force-free measurements of the conformations of DNA molecules tethered to a wall. Phys. Rev. E 83, 011916 (2011).
Ueno, H. et al. Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 98, 2014–2023 (2010).
Dunn, A.R., Chuan, P., Bryant, Z. & Spudich, J.A. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc. Natl. Acad. Sci. USA 107, 7746–7750 (2010).
Braslavsky, I. et al. Objective-type dark-field illumination for scattering from microbeads. Appl. Opt. 40, 5650–5657 (2001).
Mashanov, G.I., Tacon, D., Knight, A.E., Peckham, M. & Molloy, J.E. Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy. Methods 29, 142–152 (2003).
Friedman, L.J., Chung, J. & Gelles, J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 91, 1023–1031 (2006).
Wong, W.P. & Halvorsen, K. The effect of integration time on fluctuation measurements: calibrating an optical trap in the presence of motion blur. Opt. Express 14, 12517–12531 (2006).
Yasuda, R., Miyata, H. & Kinosita, K. Jr. Direct measurement of the torsional rigidity of single actin filaments. J. Mol. Biol. 263, 227–236 (1996).
Gore, J. et al. DNA overwinds when stretched. Nature 442, 836–839 (2006).
Lionnet, T., Joubaud, S., Lavery, R., Bensimon, D. & Croquette, V. Wringing out DNA. Phys. Rev. Lett. 96, 178102 (2006).
Sheinin, M.Y. & Wang, M.D. Twist-stretch coupling and phase transition during DNA supercoiling. Phys. Chem. Chem. Phys. 11, 4800–4803 (2009).
Heddle, J.G., Mitelheiser, S., Maxwell, A. & Thomson, N.H. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 337, 597–610 (2004).
Gutierrez-Medina, B., Andreasson, J.O., Greenleaf, W.J., Laporta, A. & Block, S.M. An optical apparatus for rotation and trapping. Methods Enzymol. 475, 377–404 (2010).
La Porta, A. & Wang, M.D. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Phys. Rev. Lett. 92, 190801 (2004).
Kauert, D.J., Kurth, T., Liedl, T. & Seidel, R. Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett. 11, 5558–5563 (2011).
Pfitzner, E. et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew. Chem. Int. Edn Engl. 52, 7766–7771 (2013).
Comstock, M.J., Ha, T. & Chemla, Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods 8, 335–340 (2011).
Huang, B., Jones, S.A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 5, 1047–1052 (2008).
Lipfert, J., Hao, X. & Dekker, N.H. Quantitative modeling and optimization of magnetic tweezers. Biophys. J. 96, 5040–5049 (2009).
Tretter, E.M. & Berger, J.M. Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J. Biol. Chem. 287, 18636–18644 (2012).
Rice, S. Mathematical analysis of random noise. Bell Syst. Tech. J. 24, 46–156 (1945).
Strick, T.R., Allemand, J.F., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996).
te Velthuis, A.J., Kerssemakers, J.W., Lipfert, J. & Dekker, N.H. Quantitative guidelines for force calibration through spectral analysis of magnetic tweezers data. Biophys. J. 99, 1292–1302 (2010).
Gosse, C. & Croquette, V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 82, 3314–3329 (2002).
Klaue, D. & Seidel, R. Torsional stiffness of single superparamagnetic microspheres in an external magnetic field. Phys. Rev. Lett. 102, 028302 (2009).
Acknowledgements
We thank J.M. Berger and members of the Bryant group for many useful discussions and comments on the manuscript, and A. Bekshaev, P. Ruijgrok and M. Dijk for providing Matlab code used for computing Mie scattering parameters and optical forces. This work was supported by a Stanford Interdisciplinary Graduate Fellowship and the Natural Sciences and Engineering Research Council of Canada (award NSERC PGS-D3) to P.L., by a Stanford Bio-X graduate fellowship to A.B., by a Pew Scholars Award and US National Institutes of Health grants OD004690 and GM106159 to Z.B., and by a Swiss National Science Foundation Fellowship to F.C.O.
Author information
Authors and Affiliations
Contributions
P.L. designed and built instrumentation, wrote data acquisition code, performed experiments and analyzed data. A.B. aided in developing and interpreting DNA gyrase experiments, collaborated on data acquisition code and performed calibration experiments to establish evanescent nanometry procedures. F.C.O. synthesized molecules for torque spectroscopy and aided in static torque assay development. E.M.T. purified and characterized E. coli GyrA and GyrB subunits for gyrase experiments. Z.B. conceived and supervised the project. P.L. and Z.B. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–3 and Supplementary Notes 1–3 (PDF 9057 kb)
High-speed darkfield imaging and angle tracking.
An 80-nm gold rotor bead was attached above a 420 bp torsionally constrained DNA segment (Supplementary Table 3 “bottom constrained” tether), held under 26 pN of tension and imaged at 6.3 kHz. A total of ∼150 ms of raw images (pixel size, 104.5 nm) are shown alongside cumulative plots of the fitted x-y positions and the corresponding calculated angle. The elapsed time in milliseconds is displayed above the image panel. x-y markers are color-coded in time to distinguish current positions from accumulated data. (MOV 2863 kb)
Comparison of predicted and observed torque-measurement distributions.
The binned data in Figure 5d are compared with the model prediction in Figure 5e by displaying torque measurement distributions –log(P(τ)) for successive values of imposed twist θ. Predicted distributions were calculated (equations S9–S12) using parameters obtained from fitting the mean torque 〈τ〉(θ) alone. The distribution is unimodal at low twist values that favor B-DNA. At intermediate twist values, the distribution becomes bimodal as formation of a Z-DNA domain becomes thermodynamically accessible; the B-Z coexistence regime is characterized by a broad torque distribution, which narrows asymmetrically as the B-Z transformation approaches completion. (MOV 5140 kb)
Supplementary Software
AuRBT data acquisition in Matlab. These files may be used to implement the data acquisition and PSF fitting procedure described in Online Methods. “Initialize.m” initializes hardware, and “Acquire.m” performs acquisition and x-y feedback. “gsolve2d.c” should be compiled as a Matlab executable, which is called by “Acquire.m” for Gaussian fitting. (ZIP 11 kb)
Rights and permissions
About this article
Cite this article
Lebel, P., Basu, A., Oberstrass, F. et al. Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nat Methods 11, 456–462 (2014). https://doi.org/10.1038/nmeth.2854
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2854
This article is cited by
-
The energy landscape for R-loop formation by the CRISPR–Cas Cascade complex
Nature Structural & Molecular Biology (2023)
-
A review on particle assembly in standing wave acoustic field
Journal of Nanoparticle Research (2022)
-
Scattering imaging of biomolecules with metallic nanoparticles: localization precision, imaging speed, and multicolor imaging capability
Optical Review (2022)
-
Optical control of fast and processive engineered myosins in vitro and in living cells
Nature Chemical Biology (2021)
-
Optical tweezers in single-molecule biophysics
Nature Reviews Methods Primers (2021)