Abstract
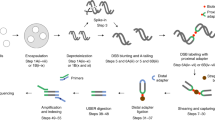
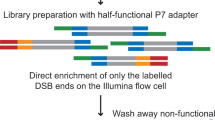
We present a genome-wide approach to map DNA double-strand breaks (DSBs) at nucleotide resolution by a method we termed BLESS (direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing). We validated and tested BLESS using human and mouse cells and different DSBs-inducing agents and sequencing platforms. BLESS was able to detect telomere ends, Sce endonuclease–induced DSBs and complex genome-wide DSB landscapes. As a proof of principle, we characterized the genomic landscape of sensitivity to replication stress in human cells, and we identified >2,000 nonuniformly distributed aphidicolin-sensitive regions (ASRs) overrepresented in genes and enriched in satellite repeats. ASRs were also enriched in regions rearranged in human cancers, with many cancer-associated genes exhibiting high sensitivity to replication stress. Our method is suitable for genome-wide mapping of DSBs in various cells and experimental conditions, with a specificity and resolution unachievable by current techniques.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Paigen, K. & Petkov, P. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11, 221–233 (2010).
Jackson, S.P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 (2010).
Branzei, D. & Foiani, M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17, 568–575 (2005).
Szilard, R.K. et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 17, 299–305 (2010).
Harrigan, J.A. et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 193, 97–108 (2011).
Seo, J. et al. Genome-wide profiles of H2AX and -H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic Acids Res. 40, 5965–5974 (2012).
Marti, T.M., Hefner, E., Feeney, L., Natale, V. & Cleaver, J.E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 103, 9891–9896 (2006).
Tuduri, S. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009).
Chadwick, B.P. & Lane, T.F. BRCA1 associates with the inactive X chromosome in late S-phase, coupled with transient H2AX phosphorylation. Chromosoma 114, 432–439 (2005).
Iacovoni, J.S. et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 (2010).
Bonner, W.M. et al. γH2AX and cancer. Nat. Rev. Cancer 8, 957–967 (2008).
Bunting, S.F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Bothmer, A. et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207, 855–865 (2010).
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 (2012).
Blitzblau, H.G., Bell, G.W., Rodriguez, J., Bell, S.P. & Hochwagen, A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 17, 2003–2012 (2007).
Feng, W. et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8, 148–155 (2006).
Feng, W., Bachant, J., Collingwood, D., Raghuraman, M.K. & Brewer, B.J. Centromere replication timing determines different forms of genomic instability in Saccharomyces cerevisiae checkpoint mutants during replication stress. Genetics 183, 1249–1260 (2009).
Leduc, F. et al. Genome-wide mapping of DNA strand breaks. PLoS ONE 6, e17353 (2011).
Dudley, D.D., Chaudhuri, J., Bassing, C.H. & Alt, F.W. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 86, 43–112 (2005).
Sfeir, A.J., Chai, W., Shay, J.W. & Wright, W.E. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell 18, 131–138 (2005).
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
Casper, A.M., Nghiem, P., Arlt, M.F. & Glover, T.W. ATR regulates fragile site stability. Cell 111, 779–789 (2002).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57, 289–300 (1995).
Smit, A. & Hubley, R. RepeatMasker Open v.3.0 〈http://www.repeatmasker.org/〉 (Institute for Systems Biology, Seattle, 1996–2004).
Durkin, S.G. & Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 (2007).
Zhang, H. & Freudenreich, C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell 27, 367–379 (2007).
Kim, N. & Jinks-Robertson, S. Transcription as a source of genome instability. Nat. Rev. Genet. 13, 204–214 (2012).
Helmrich, A., Ballarino, M. & Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011).
Halazonetis, T.D., Gorgoulis, V.G. & Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008).
Negrini, S., Gorgoulis, V.G. & Halazonetis, T.D. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010).
De, S. & Michor, F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 29, 1103–1108 (2011).
Futreal, P.A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Santarius, T., Shipley, J., Brewer, D., Stratton, M.R. & Cooper, C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10, 59–64 (2010).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Altshuler, D. et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407, 513–516 (2000).
Chiarle, R. et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011).
Crosetto, N. et al. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J. Biol. Chem. 283, 35173–35185 (2008).
Tyteca, S., Vandromme, M., Legube, G., Chevillard-Briet, M. & Trouche, D. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 25, 1680–1689 (2006).
Schmittgen, T.D. & Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Fujita, P.A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, D876–D882 (2011).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Acknowledgements
We acknowledge Y. Shiloh (Tel Aviv University) and A.J. Pierce (University of Kentucky) for kindly providing U2OS_DRH-1 cells and I-Sce plasmids. We are grateful to T. Włodarski, A.R. Lehmann, G. Fudenberg and A. Kudlicki for insightful discussions, critical reading of the manuscript and help with data analysis. This work was supported by grants from Deutsche Forschungsgemeinschaft, the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115), the LOEWE-funded Oncogenic Signaling Frankfurt network, the LOEWE Gene and Cell Therapy Center and the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013) and ERC grant agreement 250241-LineUb to I.D.; from the Associazione Italiana per la Ricerca sul cancro (AIRC) and the International Association for Cancer Research (AICR) and grant FP7 ERC-2009- StG (proposal 242965–“Lunely”) to R.C.; from the Foundation for Polish Science (TEAM), Polish National Science Centre (2011/02/A/NZ2/00014) and European Regional Development Fund under the Innovative Economy Programme (POIG.02.02.00-14-024/08-00) to K.G.; from Ligue contre le Cancer (équipe labellisée), Agence Nationale de la Recherche (RepliCare) and Institut National du Cancer to P.P.; and by grant UL1TR000071 ITS “Novel Methods” from the National Center for Research Resources, US National Institutes of Health, to M.R. M.B. is a recipient of a Human Frontier Science Program Long-Term Fellowship.
Author information
Authors and Affiliations
Contributions
N.C. and I.D. conceived and developed BLESS, coordinated the project and wrote the manuscript. A.M. developed all necessary code and analyzed Illumina data. M.J.S. and P.P. performed ChIP experiments and analysis. M.B. performed microscopy experiments and prepared figures. Q.W., E.K. and R.C. performed Roche 454 experiments and analyzed the data. N.D. contributed to statistical data analysis. M.S. and K.G. performed paired-end Illumina sequencing. M.R. conceived procedures for computational analysis, supervised the analysis and coordinated the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–7 (PDF 5109 kb)
Rights and permissions
About this article
Cite this article
Crosetto, N., Mitra, A., Silva, M. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10, 361–365 (2013). https://doi.org/10.1038/nmeth.2408
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2408
This article is cited by
-
SSBlazer: a genome-wide nucleotide-resolution model for predicting single-strand break sites
Genome Biology (2024)
-
Extru-seq: a method for predicting genome-wide Cas9 off-target sites with advantages of both cell-based and in vitro approaches
Genome Biology (2023)
-
CasKAS: direct profiling of genome-wide dCas9 and Cas9 specificity using ssDNA mapping
Genome Biology (2023)
-
A cleavage rule for selection of increased-fidelity SpCas9 variants with high efficiency and no detectable off-targets
Nature Communications (2023)
-
Discovering CRISPR–Cas off-target breaks
Nature Methods (2023)