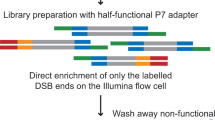
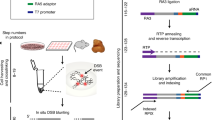
Abstract
DNA double-strand breaks (DSBs) are implicated in various physiological processes, such as class-switch recombination or crossing-over during meiosis, but also present a threat to genome stability. Extensive evidence shows that DSBs are a primary source of chromosome translocations or deletions, making them a major cause of genomic instability, a driving force of many diseases of civilization, such as cancer. Therefore, there is a great need for a precise, sensitive, and universal method for DSB detection, to enable both the study of their mechanisms of formation and repair as well as to explore their therapeutic potential. We provide a detailed protocol for our recently developed ultrasensitive and genome-wide DSB detection method: immobilized direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing (i-BLESS), which relies on the encapsulation of cells in agarose beads and labeling breaks directly and specifically with biotinylated linkers. i-BLESS labels DSBs with single-nucleotide resolution, allows detection of ultrarare breaks, takes 5 d to complete, and can be applied to samples from any organism, as long as a sufficient amount of starting material can be obtained. We also describe how to combine i-BLESS with our qDSB-Seq approach to enable the measurement of absolute DSB frequencies per cell and their precise genomic coordinates at the same time. Such normalization using qDSB-Seq is especially useful for the evaluation of spontaneous DSB levels and the estimation of DNA damage induced rather uniformly in the genome (e.g., by irradiation or radiomimetic chemotherapeutics).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alt, F. W. & Schwer, B. DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair 71, 158–163 (2018).
Gothe, H. J., Minneker, V. & Roukos, V. Dynamics of double-strand breaks: implications for the formation of chromosome translocations. Adv. Exp. Med. Biol. 1044, 27–38 (2018).
Mitelman, F., Johansson, B. & Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007).
O’Driscoll, M. Diseases associated with defective responses to DNA damage. Cold Spring Harb. Perspect. Biol. 4, a012773 (2012).
White, R. R. & Vijg, J. Do DNA double-strand breaks drive aging? Mol. Cell 63, 729–738 (2016).
Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, 1911 (2018).
Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 (2011).
Ma, A. C., Chen, Y., Blackburn, P. R. & Ekker, S. C. TALEN-mediated mutagenesis and genome editing. Methods Mol. Biol. 1451, 17–30 (2016).
Tian, X. et al. CRISPR/Cas9—An evolving biological tool kit for cancer biology and oncology. NPJ Precis. Oncol. 3, 8 (2019).
Martin, S. A., Lord, C. J. & Ashworth, A. DNA repair deficiency as a therapeutic target in cancer. Curr. Opin. Genet. Dev. 18, 80–86 (2008).
Turinetto, V. & Giachino, C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 43, 2489–2498 (2015).
McManus, K. J. & Hendzel, M. J. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell 16, 5013–5025 (2005).
Wang, F. & Higgins, J. M. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 23, 175–184 (2013).
Leduc, F. et al. Genome-wide mapping of DNA strand breaks. PLoS ONE 6, e17353 (2011).
Baranello, L. et al. DNA break mapping reveals topoisomerase II activity genome-wide. Int. J. Mol. Sci. 15, 13111–13122 (2014).
Shastri, N. et al. Genome-wide identification of structure-forming repeats as principal sites of fork collapse upon ATR inhibition. Mol. Cell 72, 222–238 e211 (2018).
Gittens, W. H. et al. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 10, 4846 (2019).
Pratto, F. et al. DNA recombination. Recombination initiation maps of individual human genomes. Science 346, 1256442 (2014).
Khil, P. P., Smagulova, F., Brick, K. M., Camerini-Otero, R. D. & Petukhova, G. V. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 22, 957–965 (2012).
Biernacka, A. et al. i-BLESS is an ultra-sensitive method for detection of DNA double-strand breaks. Commun. Biol. 1, 181 (2018).
Hoffman, E. A., McCulley, A., Haarer, B., Arnak, R. & Feng, W. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 25, 402–412 (2015).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Aymard, F. et al. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 24, 353–361 (2017).
Shi, W. et al. Ssb1 and Ssb2 cooperate to regulate mouse hematopoietic stem and progenitor cells by resolving replicative stress. Blood 129, 2479–2492 (2017).
Hu, J. et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 11, 853–871 (2016).
Lensing, S. V. et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Methods 13, 855–857 (2016).
Canela, A. et al. DNA breaks and end resection measured genome-wide by end sequencing. Mol. Cell 63, 898–911 (2016).
Mimitou, E. P., Yamada, S. & Keeney, S. A global view of meiotic double-strand break end resection. Science 355, 40–45 (2017).
Yamada, S. et al. Molecular structures and mechanisms of DNA break processing in mouse meiosis. Genes Dev. 34, 806–818 (2020).
Wiegand, R. C., Godson, G. N. & Radding, C. M. Specificity of the S1 nuclease from Aspergillus oryzae. J. Biol. Chem. 250, 8848–8855 (1975).
Yan, W. X. et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017).
Promonet, A. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11, 3940 (2020).
Zhu, Y. et al. qDSB-Seq is a general method for genome-wide quantification of DNA double-strand breaks using sequencing. Nat. Commun. 10, 2313 (2019).
Petek, L. M., Russell, D. W. & Miller, D. G. Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 18, 983–986 (2010).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Botstein, D., Chervitz, S. A. & Cherry, J. M. Yeast as a model organism. Science 277, 1259–1260 (1997).
Bassett, D. E. Jr., Boguski, M. S. & Hieter, P. Yeast genes and human disease. Nature 379, 589–590 (1996).
Zhu, Y. et al. Integrated analysis of patterns of DNA breaks reveals break formation mechanisms and their population distribution during replication stress. Preprint at https://www.biorxiv.org/content/10.1101/171439v2 (2017).
Sriramachandran, A. M. et al. Genome-wide nucleotide-resolution mapping of DNA replication patterns, single-strand breaks, and lesions by GLOE-Seq. Mol. Cell 78, 975–985 e977 (2020).
Chung, W. H., Zhu, Z., Papusha, A., Malkova, A. & Ira, G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6, e1000948 (2010).
Symington, L. S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 (2016).
Zhou, Y., Caron, P., Legube, G. & Paull, T. T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42, e19 (2014).
Aymard, F. et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014).
Vieira Braga, F. A. & Miragaia, R. J. Tissue handling and dissociation for single-cell RNA-Seq. Methods Mol. Biol. 1979, 9–21 (2019).
Reichard, A. & Asosingh, K. Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytom. A 95, 219–226 (2019).
Leelatian, N. et al. Preparing viable single cells from human tissue and tumors for cytomic analysis. Curr. Protoc. Mol. Biol. 118, 25C 21 21–25C 21 23 (2017).
Cannan, W. J. & Pederson, D. S. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell Physiol. 231, 3–14 (2016).
Li, F. et al. Apn2 resolves blocked 3′ ends and suppresses Top1-induced mutagenesis at genomic rNMP sites. Nat. Struct. Mol. Biol. 26, 155–163 (2019).
Mitra, A., Skrzypczak, M., Ginalski, K. & Rowicka, M. Strategies for achieving high sequencing accuracy for low diversity samples and avoiding sample bleeding using illumina platform. PLoS ONE 10, e0120520 (2015).
Mitra, A. et al. Analyzing and interpreting DNA double-strand break sequencing data. Preprint at https://www.biorxiv.org/content/10.1101/2020.03.05.977801v1 (2020).
Tiwari, S., Wang, S., Hagen, G. & Guilfoyle, T. J. Transfection assays with protoplasts containing integrated reporter genes. Methods Mol. Biol. 323, 237–244 (2006).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21 29 21–21 29 29 (2015).
Acknowledgements
We thank K. Jodkowska and T. Biernacki for help with the preparation of Supplementary Video 1. We also thank A. Kudlicki for critical reading of the manuscript. This work was supported by the Foundation for Polish Science (TEAM to K.G.) and the Polish National Science Centre (2015/17/D/NZ2/03711 to M.S.) and an NIH grant R01GM112131 to M.R.
Author information
Authors and Affiliations
Contributions
K.G. supervised the study, A.B., M.S., P.P., M.R., and K.G. designed the experiments, A.B. and M.S. performed the experiments, Y.Z. and M.R. performed bioinformatic analysis, A.B., M.S., Y.Z., P.P., M.R., and K.G. analyzed the results, and A.B. and K.G. wrote the manuscript. All authors read and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Anna Malkova and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Biernacka, A. et al. Commun. Biol. 1, 181 (2018): https://doi.org/10.1038/s42003-018-0165-9
Zhu, Y. et al. Nat. Commun. 10, 2313 (2019): https://doi.org/10.1038/s41467-019-10332-8
Promonet, A. et al. Nat. Commun. 11, 3940 (2020): https://doi.org/10.1038/s41467-020-17858-2
Supplementary information
Supplementary Video 1
Encapsulation of human GM19239 cells in agarose beads.
Rights and permissions
About this article
Cite this article
Biernacka, A., Skrzypczak, M., Zhu, Y. et al. High-resolution, ultrasensitive and quantitative DNA double-strand break labeling in eukaryotic cells using i-BLESS. Nat Protoc 16, 1034–1061 (2021). https://doi.org/10.1038/s41596-020-00448-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00448-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.