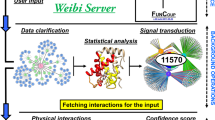
Abstract
Next-generation sequencing has not been applied to protein-protein interactome network mapping so far because the association between the members of each interacting pair would not be maintained in en masse sequencing. We describe a massively parallel interactome-mapping pipeline, Stitch-seq, that combines PCR stitching with next-generation sequencing and used it to generate a new human interactome dataset. Stitch-seq is applicable to various interaction assays and should help expand interactome network mapping.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Venkatesan, K. et al. Nat. Methods 6, 83–90 (2009).
Yu, H. et al. Science 322, 104–110 (2008).
Deplancke, B., Dupuy, D., Vidal, M. & Walhout, A.J. Genome Res. 14, 2093–2101 (2004).
Walhout, A.J. & Vidal, M. Methods 24, 297–306 (2001).
Hook, B., Bernstein, D., Zhang, B. & Wickens, M. RNA 11, 227–233 (2005).
Bennett, S.T. et al. Pharmacogenomics 6, 373–382 (2005).
Margulies, M. et al. Nature 437, 376–380 (2005).
Shendure, J. et al. Science 309, 1728–1732 (2005).
Tong, A.H. et al. Science 294, 2364–2368 (2001).
Walhout, A.J. et al. Science 287, 116–122 (2000).
Nirantar, S.R. & Ghadessy, F.J. Proteomics 11, 1335–1339 (2011).
Rual, J.F. et al. Nature 437, 1173–1178 (2005).
Lamesch, P. et al. Genomics 89, 307–315 (2007).
Braun, P. et al. Nat. Methods 6, 91–97 (2009).
Coombs, A. Nat. Biotechnol. 26, 1109–1112 (2008).
Maher, C.A. et al. Proc. Natl. Acad. Sci. USA 106, 12353–12358 (2009).
Dreze, M. et al. Methods Enzymol. 470, 281–315 (2010).
Walhout, A.J. et al. Methods Enzymol. 328, 575–592 (2000).
Walhout, A.J. & Vidal, M. Genome Res. 9, 1128–1134 (1999).
Ewing, B., Hillier, L., Wendl, M.C. & Green, P. Genome Res. 8, 175–185 (1998).
Acknowledgements
We thank members of the Dana-Farber Cancer Institute Center for Cancer Systems Biology and the Roth Laboratory for helpful discussions, and members of the University of Pennsylvania DNA Sequencing Facility in the Department of Genetics for the 454 FLX sequencing. This work was funded in part by grants R01 HG001715 (to D.E.H., F.P.R. and M.V.) and R21 HG004756 (to F.P.R.) from the US National Human Genome Research Institute of the National Institutes of Health, a grant from the Ellison Foundation (to M.V.), a Canadian Institute for Advanced Research Fellowship and Canada Excellence Research Chair (to F.P.R.) and in part by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative in support of Center for Cancer Systems Biology. M.V. is supported as a Chercheur Qualifié Honoraire from the Fonds de la Recherche Scientifique (French Community of Belgium).
Author information
Authors and Affiliations
Contributions
M.V. conceived of the project and the PCR-stitching methodology for next-generation sequencing and oversaw all aspects, including writing and editing of the manuscript; H.Y. developed the PCR-stitching process, did validations, analyzed data and co-wrote the manuscript; P.B. oversaw human interactome screening and co-wrote the manuscript; F.P.R. oversaw aspects of computational work, helped develop the next-generation sequencing strategy and contributed to editing the manuscript; D.E.H. helped with developing experimental protocols and editing the manuscript; T.H. handled database implementation and analysis of raw sequence data; L.T., S.T., E.W., F.G., N.S., T.H.-K. and J.S. carried out experimental processes for interactome mapping, PCR-stitching and validation tests; X.Y. carried out processing of PCR products and subsequent 454 FLX sequencing; C.F. and E.R. did computational and network graphing tasks; K.S.-A. contributed to troubleshooting the PCR-stitching methods; and M.E.C. contributed to writing and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–2, Supplementary Notes 1–7 (PDF 864 kb)
Rights and permissions
About this article
Cite this article
Yu, H., Tardivo, L., Tam, S. et al. Next-generation sequencing to generate interactome datasets. Nat Methods 8, 478–480 (2011). https://doi.org/10.1038/nmeth.1597
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1597
This article is cited by
-
ICDM-GEHC: identifying cancer driver module based on graph embedding and hierarchical clustering
Complex & Intelligent Systems (2024)
-
Computational biology-based study of the molecular mechanism of spermidine amelioration of acute pancreatitis
Molecular Diversity (2023)
-
Adtrp regulates thermogenic activity of adipose tissue via mediating the secretion of S100b
Cellular and Molecular Life Sciences (2022)
-
Role of FRG1 in predicting the overall survivability in cancers using multivariate based optimal model
Scientific Reports (2021)
-
Removing auto-activators from yeast-two-hybrid assays by conditional negative selection
Scientific Reports (2021)