Abstract
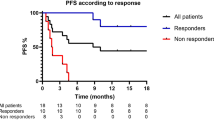
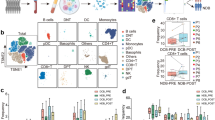
Immune-checkpoint blockade has revolutionized cancer therapy. In particular, inhibition of programmed cell death protein 1 (PD-1) has been found to be effective for the treatment of metastatic melanoma and other cancers. Despite a dramatic increase in progression-free survival, a large proportion of patients do not show durable responses. Therefore, predictive biomarkers of a clinical response are urgently needed. Here we used high-dimensional single-cell mass cytometry and a bioinformatics pipeline for the in-depth characterization of the immune cell subsets in the peripheral blood of patients with stage IV melanoma before and after 12 weeks of anti-PD-1 immunotherapy. During therapy, we observed a clear response to immunotherapy in the T cell compartment. However, before commencing therapy, a strong predictor of progression-free and overall survival in response to anti-PD-1 immunotherapy was the frequency of CD14+CD16−HLA-DRhi monocytes. We confirmed this by conventional flow cytometry in an independent, blinded validation cohort, and we propose that the frequency of monocytes in PBMCs may serve in clinical decision support.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
02 July 2018
In the version of this article initially published, Figs. 5a,c and 6a were incorrect because of an error in a metadata spreadsheet that led to the healthy donor patient 2 (HD2) samples being used twice in the analysis of baseline samples and in the analysis at 12 weeks of anti-PD-1 therapy, while HD3 samples had not been used. Data from sets of both samples should have been used in the analyses. The influence of this error on population proportions, marker expression, P values and tSNE visualization (in Figs. 5a,c and 6a) was minimal (e.g., via clustering and variance calculations). Complete reanalysis of the dataset, now including data from both HD2 and HD3, resulted in minor changes to Figs. 5a,c and 6a as well as Supplementary Figs. 7, 9, 11 and 17. This error did not affect other analyses or any of the conclusions in the paper. Also, there was an error in the description of the n values in the original Fig. 6b legend. The legend originally read: "(R and NR, n = 4 for each group)". It should be: "(HD, n = 4; R and NR, n = 3 for each group)". In addition, the Fig. 6 legend originally read: "All patients in this study were analyzed (n = 51)". It should read: "All patients from the validation cohort were analyzed (n = 31), with any patient not making the 12-month endpoint (i.e., OS or follow-up) included in the 'No' column". Additionally, there were errors in the Supplementary Information. In the ‘Validation by flow cytometry’ section of the Supplementary Methods, the antibody list was missing some antibodies. It originally read: “CD11b-BrilliantViolett (BV) 421 (ICRF44), CD14-PE (HCD14), HLA-DR-FITC (L243), CD4-BV711 (OKT4), CD33-BV605 (WM53), and Live/dead-stain-NearInfraRed." This list should include CD19-BV605 (1D3) and CD3-BV785 (OKT3). In the "Correlation of PFS with monocyte frequency" section of the Supplementary Methods, there was an error in the sentence, "To compute the cumulative hazard function we used the previously calculated cutoff of 19.39% to create the 2 groups". The percentage was incorrect. It should be 19.38%. The errors have been corrected in the HTML and PDF versions of the article.
References
Topalian, S.L., Drake, C.G. & Pardoll, D.M. Targeting the PD-1/B7-H1(PD-L1) pathway to activate antitumor immunity. Curr. Opin. Immunol. 24, 207–212 (2012).
Topalian, S.L. et al. Safety, activity and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Powles, T. et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014).
Brahmer, J. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015).
Motzer, R.J. et al. Nivolumab versus everolimus in advanced renal cell carcinoma. N. Engl. J. Med. 373, 1803–1813 (2015).
Rizvi, N.A. et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 16, 257–265 (2015).
Ansell, S.M. et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N. Engl. J. Med. 372, 311–319 (2015).
Center for Drug Evaluation Research. Approved Drugs—Hematology/Oncology (Cancer) Approvals & Safety Notifications (US Food and Drug Administration, 2016).
Hamid, O. et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144 (2013).
Nishino, M., Ramaiya, N.H., Hatabu, H. & Hodi, F.S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 14, 655–668 (2017).
Wistuba-Hamprecht, K. et al. Establishing high-dimensional immune signatures from peripheral blood via mass cytometry in a discovery cohort of stage IV melanoma patients. J. Immunol. 198, 927–936 (2017).
Pérez-Callejo, D., Romero, A., Provencio, M. & Torrente, M. Liquid-biopsy-based biomarkers in non-small-cell lung cancer for diagnosis and treatment monitoring. Transl. Lung Cancer Res. 5, 455–465 (2016).
Levine, J.H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015).
Weber, L.M. & Robinson, M. D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A 89, 1084–1096 (2016).
Maaten, L.V.D. & Hinton, G. Visualizing data using tSNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Nowicka, M. et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 6, 748 (2017).
Huang, A.C. et al. T cell invigoration to tumor burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017).
Kamphorst, A.O. et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl. Acad. Sci. USA 114, 4993–4998 (2017).
Arvaniti, E. & Claassen, M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 8, 14825 (2017).
Brahmer, J.R. et al. Phase 1 study of single-agent anti-programmed-death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics and immunologic correlates. J. Clin. Oncol. 28, 3167–3175 (2010).
Brahmer, J.R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Weber, J.S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomized, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015).
Robert, C. et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372, 2521–2532 (2015).
Ribas, A. et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. J. Am. Med. Assoc. 315, 1600–1609 (2016).
Ostrand-Rosenberg, S. & Sinha, P. Myeloid-derived suppressor cells: linking inflammation and cancer. J. Immunol. 182, 4499–4506 (2009).
Gebhardt, C. et al. Myeloid cells and related chronic inflammatory factors as novel predictive markers in melanoma treatment with ipilimumab. Clin. Cancer Res. 21, 5453–5459 (2015).
Meyer, C. et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 63, 247–257 (2014).
Sade-Feldman, M. et al. Clinical significance of circulating CD33+CD11b+HLA-DR− myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clin. Cancer Res. 22, 5661–5672 (2016).
Komohara, Y., Jinushi, M. & Takeya, M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 105, 1–8 (2014).
Zhang, Q.-W. et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 7, e50946 (2012).
Romano, E. et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc. Natl. Acad. Sci. USA 112, 6140–6145 (2015).
Chavan, R. et al. Untreated stage IV melanoma patients exhibit abnormal monocyte phenotypes and decreased functional capacity. Cancer Immunol. Res. 2, 241–248 (2014).
Zaretsky, J.M. et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016).
Bellucci, R. et al. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. OncoImmunology 4, e1008824 (2015).
Daud, A.I. et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Invest. 126, 3447–3452 (2016).
Herbst, R.S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014).
Kluger, H.M. et al. Characterization of PD-L1 expression and associated T cell infiltrates in metastatic melanoma samples from variable anatomic sites. Clin. Cancer Res. 21, 3052–3060 (2015).
von Andrian, U.H. & Mempel, T.R. Homing and cellular traffic in lymph nodes. Nat. Rev. Immunol. 3, 867–878 (2003).
Takizawa, H., Regoes, R.R., Boddupalli, C.S., Bonhoeffer, S. & Manz, M.G. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208, 273–284 (2011).
Nagai, Y. et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 24, 801–812 (2006).
Caux, C., Moreau, I., Saeland, S. & Banchereau, J. Interferon-γ enhances factor-dependent myeloid proliferation of human CD34+ hematopoietic progenitor cells. Blood 79, 2628–2635 (1992).
Hartmann, F.J. et al. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J. Exp. Med. 213, 2621–2633 (2016).
Spidlen, J. & Brinkman, R.R. Use FlowRepository to share your clinical data upon study publication. Cytometry B Clin. Cytom. (2016).
Finck, R. et al. Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494 (2013).
Patro, R., Duggal, G., Love, M.I., Irizarry, R.A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Soneson, C., Love, M.I. & Robinson, M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D.J., Chen, Y. & Smyth, G.K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Acknowledgements
We thank V. Tosevski and T.M. Brodie (mass cytometry core facility, University of Zurich), A. Langer (Department of Dermatology, University of Zurich), and C. Beisel and K. Eschbach (Genomics Facility, ETH Basel) for excellent technical assistance and N. Nunes, B. Chatterjee, E. Terskikh, and C. Gujer (all from the Institute of Experimental Immunology, University Zurich), A. Zollinger (Swiss Institute of Bioinformatics, Lausanne), all members of the COST Action BM1404 Mye-EUNITER (http://www.mye-euniter.eu), and P. Cheng (University of Zurich) for discussions. We also thank C. Guglietta for graphical design and layout. This work received funding from the University Research Priority Program (URPP) in Translational Cancer Research (C.K.), the Swiss National Science Foundation (grants 310030_146130 and 316030_150768; B.B.), the European Union FP7 project ATECT (B.B.), and the European Training Network MELGEN (M.P.L.).
Author information
Authors and Affiliations
Contributions
C.K., M.P.L., and B.B. conceived the study and analyzed data; C.K., S.G., and B.B. designed and performed the experiments; F.J.H. and S.G. assisted with the experiments; S.S., R.D., and M.P.L. provided clinical samples and performed statistical analyses of clinical parameters; R.D. and M.P.L. analyzed histology; M.N., L.M.W., and M.D.R. provided analysis algorithms and analyzed data; C.K. and S.G. wrote the manuscript; M.P.L., M.D.R., and B.B. edited the manuscript; and all authors read and gave final approval to submit the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Methods, Supplementary Tables 1–3 & Supplementary Figures 1–17 (PDF 7906 kb)
Rights and permissions
About this article
Cite this article
Krieg, C., Nowicka, M., Guglietta, S. et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 24, 144–153 (2018). https://doi.org/10.1038/nm.4466
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4466
This article is cited by
-
SuperCellCyto: enabling efficient analysis of large scale cytometry datasets
Genome Biology (2024)
-
Translational research on drug development and biomarker discovery for hepatocellular carcinoma
Journal of Biomedical Science (2024)
-
Blood-based biomarkers in patients with non-small cell lung cancer treated with immune checkpoint blockade
Journal of Experimental & Clinical Cancer Research (2024)
-
CD137+ and regulatory T cells as independent prognostic factors of survival in advanced non-oncogene addicted NSCLC patients treated with immunotherapy as first-line
Journal of Translational Medicine (2024)
-
Dendritic cells as orchestrators of anticancer immunity and immunotherapy
Nature Reviews Clinical Oncology (2024)