Abstract
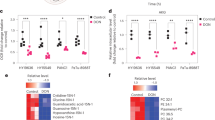
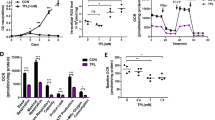
The unique metabolic demands of cancer cells underscore potentially fruitful opportunities for drug discovery in the era of precision medicine. However, therapeutic targeting of cancer metabolism has led to surprisingly few new drugs to date. The neutral amino acid glutamine serves as a key intermediate in numerous metabolic processes leveraged by cancer cells, including biosynthesis, cell signaling, and oxidative protection. Herein we report the preclinical development of V-9302, a competitive small molecule antagonist of transmembrane glutamine flux that selectively and potently targets the amino acid transporter ASCT2. Pharmacological blockade of ASCT2 with V-9302 resulted in attenuated cancer cell growth and proliferation, increased cell death, and increased oxidative stress, which collectively contributed to antitumor responses in vitro and in vivo. This is the first study, to our knowledge, to demonstrate the utility of a pharmacological inhibitor of glutamine transport in oncology, representing a new class of targeted therapy and laying a framework for paradigm-shifting therapies targeting cancer cell metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pochini, L., Scalise, M., Galluccio, M. & Indiveri, C. Membrane transporters for the special amino acid glutamine: structure/function relationships and relevance to human health. Front. Chem. 2, 61 (2014).
Jin, L., Alesi, G.N. & Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 35, 3619–3625 (2016).
Hassanein, M. et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin. Cancer Res. 19, 560–570 (2013).
van Geldermalsen, M. et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35, 3201–3208 (2016).
Schulte, M.L. et al. Non-invasive glutamine PET reflects pharmacological inhibition of BRAFV600Ein vivo. Mol. Imaging Biol. 19, 421–428 (2017).
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009).
Watanabe, T. et al. Differential gene expression signatures between colorectal cancers with and without KRAS mutations: crosstalk between the KRAS pathway and other signalling pathways. Eur. J. Cancer 47, 1946–1954 (2011).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017).
Shukla, K. et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-henylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 55, 10551–10563 (2012).
Harding, J.J. et al. Safety and tolerability of increasing doses of CB-839, a first-in-class, orally administered small molecule inhibitor of glutaminase, in solid tumors. J. Clin. Oncol. 33, 2512 (2015).
Rhoads, J.M. et al. Glutamine metabolism stimulates intestinal cell MAPKs by a cAMP-inhibitable, Raf-independent mechanism. Gastroenterology 118, 90–100 (2000).
Willems, L. et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 122, 3521–3532 (2013).
Schulte, M.L., Khodadadi, A.B., Cuthbertson, M.L., Smith, J.A. & Manning, H.C. 2-Amino-4-bis(aryloxybenzyl)aminobutanoic acids: a novel scaffold for inhibition of ASCT2-mediated glutamine transport. Bioorg. Med. Chem. Lett. 26, 1044–1047 (2016).
Esslinger, C.S., Cybulski, K.A. & Rhoderick, J.F. Nγ-Aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg. Med. Chem. 13, 1111–1118 (2005).
Lomenick, B. et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106, 21984–21989 (2009).
Canul-Tec, J.C. et al. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544, 446–451 (2017).
Fuchs, B.C. & Bode, B.P. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 15, 254–266 (2005).
Nicklin, P. et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009).
Vichai, V. & Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 1, 1112–1116 (2006).
Rathmell, J.C. T cell Myc-tabolism. Immunity 35, 845–846 (2011).
Skala, M. & Ramanujam, N. Multiphoton redox ratio imaging for metabolic monitoring in vivo. Methods Mol. Biol. 594, 155–162 (2010).
Walsh, A.J. et al. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 74, 5184–5194 (2014).
DeBerardinis, R.J., Sayed, N., Ditsworth, D. & Thompson, C.B. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 (2008).
Hanover, J.A., Krause, M.W. & Love, D.C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800, 80–95 (2010).
Obeid, L.M., Linardic, C.M., Karolak, L.A. & Hannun, Y.A. Programmed cell death induced by ceramide. Science 259, 1769–1771 (1993).
Tresse, E., Kosta, A., Giusti, C., Luciani, M.F. & Golstein, P. A UDP-glucose derivative is required for vacuolar autophagic cell death. Autophagy 4, 680–691 (2008).
Sentelle, R.D. et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat. Chem. Biol. 8, 831–838 (2012).
Dall'Armi, C., Devereaux, K.A. & Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 23, R33–R45 (2013).
Shatz, O., Holland, P., Elazar, Z. & Simonsen, A. Complex relations between phospholipids, autophagy, and neutral lipids. Trends Biochem. Sci. 41, 907–923 (2016).
Huang, F., Zhang, Q., Ma, H., Lv, Q. & Zhang, T. Expression of glutaminase is upregulated in colorectal cancer and of clinical significance. Int. J. Clin. Exp. Pathol. 7, 1093–1100 (2014).
Xiang, Y. et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306 (2015).
Seltzer, M.J. et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 70, 8981–8987 (2010).
Gross, M.I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 (2014).
Suzuki, S. et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 107, 7461–7466 (2010).
Chiu, M. et al. GPNA inhibits the sodium-independent transport system L for neutral amino acids. Amino Acids 49, 1365–1372 (2017).
McKinley, E.T., Zhao, P., Coffey, R.J., Washington, M.K. & Manning, H.C. 3′-Deoxy-3′-[18F]-fluorothymidine PET imaging reflects PI3K–mTOR-mediated pro-survival response to targeted therapy in colorectal cancer. PLoS One 9, e108193 (2014).
Wiza, C., Nascimento, E.B. & Ouwens, D.M. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am. J. Physiol. Endocrinol. Metab. 302, E1453–E1460 (2012).
Santio, N.M. et al. The PIM1 kinase promotes prostate cancer cell migration and adhesion via multiple signalling pathways. Exp. Cell Res. 342, 113–124 (2016).
Rhoads, J.M. et al. L-Glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Physiol. 272, G943–G953 (1997).
Meiler, J. & Baker, D. ROSETTALIGAND: protein–small molecule docking with full side-chain flexibility. Proteins 65, 538–548 (2006).
Kondo, J. et al. Retaining cell–cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc. Natl. Acad. Sci. USA 108, 6235–6240 (2011).
McKinley, E.T. et al. 18FDG-PET predicts pharmacodynamic response to OSI-906, a dual IGF-1R/IR inhibitor, in preclinical mouse models of lung cancer. Clin. Cancer Res. 17, 3332–3340 (2011).
Acknowledgements
The authors acknowledge M. Tantawy for assistance with PET imaging, F. Revetta for histology expertise, and A. Rosenberg and A. Cohen for helpful discussions and editorial assistance. The authors wish to acknowledge research support from the Vanderbilt Ingram Cancer Center Support Grant (National Institutes of Health (NIH) National Cancer Institute (NCI) P30CA068485, H.C.M.), which supports the Vanderbilt-Ingram Cancer Center (VICC) Chemical Synthesis Core, Vanderbilt University Medical Center (VUMC) Radiochemistry Core, and Center for Small Animal Imaging; the Kleberg Foundation (H.C.M.); a Vanderbilt Trans-Institutional Program (TIPS) Award to the Vanderbilt Center for Molecular Probes (H.C.M.); the Vanderbilt Specialized Program of Research Excellence (SPORE) in Gastrointestinal Cancer (NIH NCI P50CA095103, R.J.C. and H.C.M.); an Outstanding Investigator Award from the NCI (R35CA197570, R.J.C.); and the Vanderbilt Digestive Disease Research Center (NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) P30DK058404, H.C.M.).
Author information
Authors and Affiliations
Contributions
M.L.S. performed or designed most of the experiments with assistance from A.F., L.G., P.Z., and J.L. S.T.S. and J.A.S. performed computational modeling. J.K. and R.J.C. performed organoid studies. J.B. enrolled individuals who provided tissues for organoids and PDXs. J.T.S. and M.C.S. performed optical redox ratio measurements. M.O.J. and J.C.R. performed T cell experiments. M.K.W. interpreted pathology samples. M.L.N. oversaw radiopharmaceutical production and provided technical assistance. H.C.M. designed and supervised the study. M.L.S., M.L.N., and H.C.M. wrote the manuscript. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21 (PDF 4353 kb)
Supplementary Data 1
Full in vivo metabolomic data set (XLSX 627 kb)
Rights and permissions
About this article
Cite this article
Schulte, M., Fu, A., Zhao, P. et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med 24, 194–202 (2018). https://doi.org/10.1038/nm.4464
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4464
This article is cited by
-
Glutamine addiction in tumor cell: oncogene regulation and clinical treatment
Cell Communication and Signaling (2024)
-
A glutamine tug-of-war between cancer and immune cells: recent advances in unraveling the ongoing battle
Journal of Experimental & Clinical Cancer Research (2024)
-
The pleiotropic functions of reactive oxygen species in cancer
Nature Cancer (2024)
-
Crosstalk between autophagy and metabolism: implications for cell survival in acute myeloid leukemia
Cell Death Discovery (2024)
-
Amino acid metabolism in tumor biology and therapy
Cell Death & Disease (2024)