Abstract
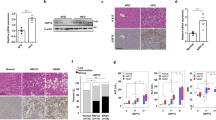
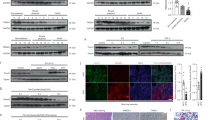
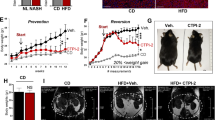
Nonalcoholic steatohepatitis (NASH) is a common clinical condition that can lead to advanced liver diseases. Lack of effective pharmacotherapies for NASH is largely attributable to an incomplete understanding of its pathogenesis. The deubiquitinase cylindromatosis (CYLD) plays key roles in inflammation and cancer. Here we identified CYLD as a suppressor of NASH in mice and in monkeys. CYLD is progressively degraded upon interaction with the E3 ligase TRIM47 in proportion to NASH severity. We observed that overexpression of Cyld in hepatocytes concomitantly inhibits lipid accumulation, insulin resistance, inflammation and fibrosis in mice with NASH induced in an experimental setting. Mechanistically, CYLD interacts directly with the kinase TAK1 and removes its K63-linked polyubiquitin chain, which blocks downstream activation of the JNK–p38 cascades. Notably, reconstitution of hepatic CYLD expression effectively reverses disease progression in mice with dietary or genetically induced NASH and in high-fat diet–fed monkeys predisposed to metabolic syndrome. Collectively, our findings demonstrate that CYLD mitigates NASH severity and identify the CYLD–TAK1 axis as a promising therapeutic target for management of the disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Byrne, C.D. & Targher, G. NAFLD: a multisystem disease. J. Hepatol. 62 (Suppl.), S47–S64 (2015).
Rinella, M.E. Nonalcoholic fatty liver disease: a systematic review. J. Am. Med. Assoc. 313, 2263–2273 (2015).
Younossi, Z.M. et al. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016).
Chalasani, N. et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology https://doi.org/10.1002/hep.29367 (2017).
Michelotti, G.A., Machado, M.V. & Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 10, 656–665 (2013).
Hardy, T., Oakley, F., Anstee, Q.M. & Day, C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu. Rev. Pathol. 11, 451–496 (2016).
Suzuki, A. & Diehl, A.M. Nonalcoholic steatohepatitis. Annu. Rev. Med. 68, 85–98 (2017).
Rotman, Y. & Sanyal, A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut 66, 180–190 (2017).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Massoumi, R. Ubiquitin chain cleavage: CYLD at work. Trends Biochem. Sci. 35, 392–399 (2010).
Yoshida, H., Jono, H., Kai, H. & Li, J.D. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for Toll-like receptor 2 signaling via negative cross-talk with TRAF6 and TRAF7. J. Biol. Chem. 280, 41111–41121 (2005).
Massoumi, R., Chmielarska, K., Hennecke, K., Pfeifer, A. & Fässler, R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125, 665–677 (2006).
Reiley, W.W. et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7, 411–417 (2006).
Hutti, J.E. et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKɛ promotes cell transformation. Mol. Cell 34, 461–472 (2009).
Wu, X. et al. SCFβ–TRCP regulates osteoclastogenesis via promoting CYLD ubiquitination. Oncotarget 5, 4211–4221 (2014).
Zhao, X. et al. Gadolinium chloride ameliorates acute lung injury associated with severe acute pancreatitis in rats by regulating CYLD/NF-κB signaling. Biochem. Biophys. Res. Commun. 492, 255–261 (2017).
Yu, B. et al. CYLD deubiquitinates nicotinamide adenine dinucleotide phosphate oxidase 4 contributing to adventitial remodeling. Arterioscler. Thromb. Vasc. Biol. 37, 1698–1709 (2017).
Lee, B.C., Miyata, M., Lim, J.H. & Li, J.D. Deubiquitinase CYLD acts as a negative regulator for bacterium NTHi-induced inflammation by suppressing K63-linked ubiquitination of MyD88. Proc. Natl. Acad. Sci. USA 113, E165–E171 (2016).
Lim, J.H. et al. CYLD negatively regulates transforming growth factor-β-signalling via deubiquitinating Akt. Nat. Commun. 3, 771 (2012).
Trompouki, E. et al. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia 11, 469–476 (2009).
Reissig, S. et al. The deubiquitinating enzyme CYLD regulates the differentiation and maturation of thymic medullary epithelial cells. Immunol. Cell Biol. 93, 558–566 (2015).
Zhao, Y. et al. CYLD and the NEMO zinc finger regulate tumor necrosis factor signaling and early embryogenesis. J. Biol. Chem. 290, 22076–22084 (2015).
Annunziata, C.M. et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Keats, J.J. et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007).
Hellerbrand, C. et al. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 28, 21–27 (2007).
Tsagaratou, A., Trompouki, E., Grammenoudi, S., Kontoyiannis, D.L. & Mosialos, G. Thymocyte-specific truncation of the deubiquitinating domain of CYLD impairs positive selection in a NF-κB essential modulator-dependent manner. J. Immunol. 185, 2032–2043 (2010).
Reissig, S. et al. The tumor suppressor CYLD controls the function of murine regulatory T cells. J. Immunol. 189, 4770–4776 (2012).
Hellerbrand, C. & Massoumi, R. Cylindromatosis—a protective molecule against liver diseases. Med. Res. Rev. 36, 342–359 (2016).
Urbanik, T. et al. Liver specific deletion of CYLDexon7/8 induces severe biliary damage, fibrosis and increases hepatocarcinogenesis in mice. J. Hepatol. 57, 995–1003 (2012).
Nikolaou, K. et al. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell 21, 738–750 (2012).
Haas, J.T., Francque, S. & Staels, B. Pathophysiology and mechanisms of nonalcoholic fatty liver disease. Annu. Rev. Physiol. 78, 181–205 (2016).
Mathis, B.J. et al. CYLD-mediated signaling and diseases. Curr. Drug Targets 16, 284–294 (2015).
Sanches, S.C., Ramalho, L.N., Augusto, M.J., da Silva, D.M. & Ramalho, F.S. Nonalcoholic steatohepatitis: a search for factual animal models. BioMed Res. Int. 2015, 574832 (2015).
Arthur, J.S. & Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13, 679–692 (2013).
Singh, A. et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 148, 639–650 (2012).
Ji, Y.-X. The ubiquitin E3 ligase TRAF6 exacerbates pathological cardiac hypertrophy via TAK1-dependent signalling. Nat. Commun. 7, 11267 (2016).
Reiley, W.W. et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204, 1475–1485 (2007).
Oseini, A.M. & Sanyal, A.J. Therapies in non-alcoholic steatohepatitis (NASH). Liver Int. 37 (Suppl 1.), 97–103 (2017).
Caldwell, S. NASH therapy: omega 3 supplementation, vitamin E, insulin sensitizers and statin drugs. Clin. Mol. Hepatol. 23, 103–108 (2017).
Ajibade, A.A., Wang, H.Y. & Wang, R.F. Cell type–specific function of TAK1 in innate immune signaling. Trends Immunol. 34, 307–316 (2013).
Dai, L., Aye Thu, C., Liu, X.Y., Xi, J. & Cheung, P.C. TAK1, more than just innate immunity. IUBMB Life 64, 825–834 (2012).
Mihaly, S.R., Ninomiya-Tsuji, J. & Morioka, S. TAK1 control of cell death. Cell Death Differ. 21, 1667–1676 (2014).
An, S. et al. USP18 protects against hepatic steatosis and insulin resistance through its deubiquitinating activity. Hepatology 66, 1866–1884 (2017).
Wang, P.X. et al. Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nat. Commun. 7, 10592 (2016).
Yan, F.J. et al. The E3 ligase tripartite motif 8 targets TAK1 to promote insulin resistance and steatohepatitis. Hepatology 65, 1492–1511 (2016).
Seki, E. TAK1-dependent autophagy: a suppressor of fatty liver disease and hepatic oncogenesis. Mol. Cell. Oncol. 1, e968507 (2014).
Roh, Y.S., Song, J. & Seki, E. TAK1 regulates hepatic cell survival and carcinogenesis. J. Gastroenterol. 49, 185–194 (2014).
Yang, L. et al. Transforming growth factor-β signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology 144, 1042–1054 e4 (2013).
Wang, C. et al. Melittin, a major component of bee venom, sensitizes human hepatocellular carcinoma cells to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)-induced apoptosis by activating CaMKII–TAK1–JNK/p38 and inhibiting IκBα kinase–NF-κB. J. Biol. Chem. 284, 3804–3813 (2009).
Zhao, N. et al. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol. Cancer 13, 35 (2014).
Tey, S.K. et al. Nuclear Met promotes hepatocellular carcinoma tumorigenesis and metastasis by upregulation of TAK1 and activation of NF-κB pathway. Cancer Lett. 411, 150–161 (2017).
Gao, L. et al. Tumor necrosis factor receptor–associated factor 5 (Traf5) acts as an essential negative regulator of hepatic steatosis. J. Hepatol. 65, 125–136 (2016).
Xiang, M. et al. Targeting hepatic TRAF1–ASK1 signaling to improve inflammation, insulin resistance, and hepatic steatosis. J. Hepatol. 64, 1365–1377 (2016).
Zhang, X.F. et al. TRAF1 is a key mediator for hepatic ischemia/reperfusion injury. Cell Death Dis. 5, e1467 (2014).
Hu, J. et al. Targeting TRAF3 signaling protects against hepatic ischemia/reperfusions injury. J. Hepatol. 64, 146–159 (2016).
Wang, P.X. et al. Targeting CASP8 and FADD-like apoptosis regulator ameliorates nonalcoholic steatohepatitis in mice and nonhuman primates. Nat. Med. 23, 439–449 (2017).
Kleiner, D.E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Zhao, G.N. et al. Tmbim1 is a multivesicular body regulator that protects against non-alcoholic fatty liver disease in mice and monkeys by targeting the lysosomal degradation of Tlr4. Nat. Med. 23, 742–752 (2017).
Acknowledgements
We thank H.-B. Shu (Medical Research Institute, Wuhan University) for providing ubiquitin and several of its derivative plasmids. We thank Y. Rao, M. Guo and R. Zhang (Institute of Model Animal of Wuhan University) for their assistance in monkey experiments. This work was supported by grants from the National Science Fund for Distinguished Young Scholars (no. 81425005; H.L.), the Key Project of the National Natural Science Foundation (no. 81330005 and 81630011; H.L.), the National Science and Technology Support Project (no. 2014BAI02B01 and 2015BAI08B01; H.L.), the National Key Research and Development Program (no. 2013YQ030923-05 (H.L.) and no. 2016YFF0101500 (Z.-G.S.)), the National Natural Science Foundation of China (no. 81770053 (Z.-G.S.) and no. 91729303 (L.C.)), the Key Collaborative Project of the National Natural Science Foundation (no. 91639304; H.L.) and the National Institutes of Health (no. DK048873, DK056626 and DK103046; D.E.C.).
Author information
Authors and Affiliations
Contributions
Y.-X.J., Z.H., X.Y. and X.W. designed and performed the experiments, analyzed data and wrote the manuscript; L.-P.Z. performed molecular experiments, analyzed data and wrote the manuscript; P.-X.W. performed animal experiments and analyzed data; X.-J.Z., M.A.-B. and L.C. wrote the manuscript and provided important advice for this study; P.Z., Y.-X.L., L.B., M.-M.G. and H.Z. performed biological experiments and analyzed data; S.T. established mouse and monkey NASH models; Y.W. and Z.-X.H. performed the experiments involving monkeys; X.-Y.Z. performed western blot experiments; Y.Z. performed histopathological analysis; J.G. constructed the genetically engineered mice and performed AAV8 construction; Z.-G.S. and F.L. wrote the manuscript and provided important advice for this study; D.E.C. and H.L. designed experiments, wrote the manuscript and supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Figures 1–12 & Supplementary Tables 1–4 (PDF 19998 kb)
Life Sciences Reporting Summary
Life Sciences Reporting Summary (PDF 219 kb)
Rights and permissions
About this article
Cite this article
Ji, YX., Huang, Z., Yang, X. et al. The deubiquitinating enzyme cylindromatosis mitigates nonalcoholic steatohepatitis. Nat Med 24, 213–223 (2018). https://doi.org/10.1038/nm.4461
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4461
This article is cited by
-
USP9X-mediated NRP1 deubiquitination promotes liver fibrosis by activating hepatic stellate cells
Cell Death & Disease (2023)
-
MALT1-dependent cleavage of CYLD promotes NF-κB signaling and growth of aggressive B-cell receptor-dependent lymphomas
Blood Cancer Journal (2023)
-
Ablation of the deubiquitinase USP15 ameliorates nonalcoholic fatty liver disease and nonalcoholic steatohepatitis
Experimental & Molecular Medicine (2023)
-
Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies
Signal Transduction and Targeted Therapy (2023)
-
Selenium-Enriched Probiotic Alleviates Western Diet-Induced Non-alcoholic Fatty Liver Disease in Rats via Modulation of Autophagy Through AMPK/SIRT-1 Pathway
Biological Trace Element Research (2023)