Abstract
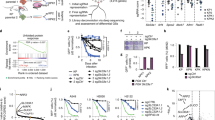
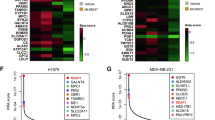
Treating KRAS-mutant lung adenocarcinoma (LUAD) remains a major challenge in cancer treatment given the difficulties associated with directly inhibiting the KRAS oncoprotein1. One approach to addressing this challenge is to define mutations that frequently co-occur with those in KRAS, which themselves may lead to therapeutic vulnerabilities in tumors. Approximately 20% of KRAS-mutant LUAD tumors carry loss-of-function mutations in the KEAP1 gene encoding Kelch-like ECH-associated protein 1 (refs. 2, 3, 4), a negative regulator of nuclear factor erythroid 2-like 2 (NFE2L2; hereafter NRF2), which is the master transcriptional regulator of the endogenous antioxidant response5,6,7,8,9,10. The high frequency of mutations in KEAP1 suggests an important role for the oxidative stress response in lung tumorigenesis. Using a CRISPR–Cas9-based approach in a mouse model of KRAS-driven LUAD, we examined the effects of Keap1 loss in lung cancer progression. We show that loss of Keap1 hyperactivates NRF2 and promotes KRAS-driven LUAD in mice. Through a combination of CRISPR–Cas9-based genetic screening and metabolomic analyses, we show that Keap1- or Nrf2-mutant cancers are dependent on increased glutaminolysis, and this property can be therapeutically exploited through the pharmacological inhibition of glutaminase. Finally, we provide a rationale for stratification of human patients with lung cancer harboring KRAS/KEAP1- or KRAS/NRF2-mutant lung tumors as likely to respond to glutaminase inhibition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Cox, A.D., Fesik, S.W., Kimmelman, A.C., Luo, J. & Der, C.J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014).
Berger, A.H. et al. High-throughput phenotyping of lung cancer somatic mutations. Cancer Cell 30, 214–228 (2016).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Singh, A. et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3, e420 (2006).
Itoh, K. et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322 (1997).
Itoh, K. et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 (1999).
Mitsuishi, Y. et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012).
Harris, I.S. et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 27, 211–222 (2015).
DeNicola, G.M. et al. NRF2 regulates serine biosynthesis in non–small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015).
Sullivan, L.B., Gui, D.Y. & Heiden, M.G.V. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat. Rev. Cancer 16, 680–693 (2016).
DuPage, M., Dooley, A.L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009).
Sánchez-Rivera, F.J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014).
Mazur, P.K. et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 21, 1163–1171 (2015).
Davidson, S.M. et al. Environment impacts the metabolic dependencies of ras-driven non–small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Maresch, R. et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 7, 10770 (2016).
McKenna, A. et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016).
Goldstein, L.D. et al. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 16, 2605–2617 (2016).
Meylan, E. et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature 462, 104–107 (2009).
Singh, A. et al. RNAi-mediated silencing of nuclear factor erythroid-2–related factor 2 gene expression in non–small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 68, 7975–7984 (2008).
Malhotra, D. et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP–seq profiling and network analysis. Nucleic Acids Res. 38, 5718–5734 (2010).
Hassanein, M. et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin. Cancer Res. 19, 560–570 (2013).
Metallo, C.M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011).
Altman, B.J., Stine, Z.E. & Dang, C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Bauer, A.K. et al. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One 6, e26590 (2011).
DeNicola, G.M. et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011).
Satoh, H., Moriguchi, T., Takai, J., Ebina, M. & Yamamoto, M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 73, 4158–4168 (2013).
Sayin, V.I. et al. Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 6, 221ra15 (2014).
Chio, I.I. et al. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166, 963–976 (2016).
Satoh, H. et al. NRF2 intensifies host defense systems to prevent lung carcinogenesis, but after tumor initiation accelerates malignant cell growth. Cancer Res. 76, 3088–3096 (2016).
Kerr, E.M., Gaude, E., Turrell, F.K., Frezza, C. & Martins, C.P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113 (2016).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Jaramillo, M.C. & Zhang, D.D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 (2013).
Konstantinopoulos, P.A. et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 71, 5081–5089 (2011).
Shibata, T. et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135, 1358–1368 (2008).
Kim, Y.R. et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 220, 446–451 (2010).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 (2013).
Fabrizio, F.P. et al. Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 8, 11187–11198 (2017).
Muscarella, L.A. et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient's outcome. Epigenetics 6, 317–325 (2011).
Hanada, N. et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12, 66 (2012).
Krall, E.B. et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 6, e18970 (2017).
Jackson, E.L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Jackson, E.L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Dimitrova, N. et al. Stromal expression of miR-143/145 promotes neoangiogenesis in lung cancer development. Cancer Discov. 6, 188–201 (2016).
Sanjana, N.E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Papagiannakopoulos, T. et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 24, 324–331 (2016).
Abouelhoda, M.I., Kurtz, S. & Ohlebusch, E. Replacing suffix trees with enhanced suffix arrays. J. Discrete Algorithms (AMST) 2, 53–86 (2004).
Smith, T.F. & Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Döring, A., Weese, D., Rausch, T. & Reinert, K. SeqAn an efficient, generic C. library for sequence analysis. BMC Bioinformatics 9, 11 (2008).
Thorvaldsdóttir, H., Robinson, J.T. & Mesirov, J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Tarasov, A., Vilella, A.J., Cuppen, E., Nijman, I.J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Gardner, E.E. et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2–SLFN1L axis. Cancer Cell 31, 286–299 (2017).
Cheng, D.T. et al. Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Schneeberger, V.E., Allaj, V., Gardner, E.E., Poirier, J.T. & Rudin, C.M. Quantitation of murine stroma and selective purification of the human tumor component of patient-derived xenografts for genomic analysis. PLoS One 11, e0160587 (2016).
Barbie, D.A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Singh, A. et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Invest. 123, 2921–2934 (2013).
Ritchie, M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Bullard, J.H., Purdom, E., Hansen, K.D. & Dudoit, S. Evaluation of statistical methods for normalization and differential expression inmRNA-Seq experiments. BMC Bioinformatics 11, 94 (2010).
Li, C.M. et al. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev. 29, 1850–1862 (2015).
Biton, A. et al. Independent component analysis uncovers the landscape of the bladder tumor transcriptome and reveals insights into luminal and basal subtypes. Cell Rep. 9, 1235–1245 (2014).
Lewis, C.A. et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014).
Young, J.D., Walther, J.L., Antoniewicz, M.R., Yoo, H. & Stephanopoulos, G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol. Bioeng. 99, 686–699 (2008).
Fernandez, C.A., Des Rosiers, C., Previs, S.F., David, F. & Brunengraber, H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 (1996).
Acknowledgements
We thank D. McFadden, R. Possemato, S. Sayin, and T. González-Robles for critical reading of the manuscript; T. Tammela, L. Sullivan, G. DeNicola, and I. Harris for scientific discussions and feedback; S. Levine and T. Mason for massively parallel sequencing expertise; M. Griffin, M. Jennings, and G. Paradis for fluorescence-activated cell sorting (FACS) support; K. Cormier and the Hope Babette Tang (1983) Histology Facility for histology support; I. Baptista, A. Deconinck, J. Teixeira, and K. Yee for administrative support; and the Swanson Biotechnology Center for excellent core facilities. This work was supported in part by the Laura and Isaac Perlmutter Cancer Support Grant, National Institutes of Health (NIH) S10 awards, and Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. T.P. was supported by the American Cancer Society and Hope Funds for Cancer Research. The laboratory of T.P. is supported by the NIH (K22CA201088-01) and the New York University Department of Pathology Bridge Grant. R.R. was supported by the National Science Foundation Graduate Research Fellowship under grant number 1122374. V.I.S. received support from the Swedish Medical Research Council, the AG Fond, and the Wenner-Gren Foundations and is the recipient of EMBO long-term fellowship ALTF 1451-2015 that is co-funded by the European Commission (LTCOFUND2013, GA-2013-609409) with support from Marie Curie Actions. S.E.L. is supported by an NIH training grant (5T32HL007151-38). Human tumor collection by H.I.P. was supported by a National Cancer Institute Early Detection Research Network grant (2U01CA 111295-04). Research in the laboratory of T.J. was supported by Cancer Center Support Grant P30-CA14051 and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
R.R., V.I.S., F.J.S.-R., T.J., and T.P. designed the study; R.R., V.I.S., M.R.B., S.M.D., S.X.S., S.E.L., T.R.K., D.C.E., L.S., and B.M. performed experiments; A.B. and I.D. conducted bioinformatic analyses; S.M.D. and M.G.V.H. provided feedback and interpretation of metabolism data; E.E.S. and J.R.P. provided custom NRF2 antibody; C.J.T. provided advice and feedback on CB-839 administration; R.T.B. performed histopathological analysis of GEMMs; A.D., V.A., J.T.P., and C.M.R. generated and characterized PDX models; I.D., A.H., A.L.M., C.G., and H.I.P. were involved in human tumor collection, sequencing, and characterization; R.R., V.I.S., T.J., and T.P. wrote the manuscript with comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12. (PDF 3721 kb)
Supplementary Table 1
Targeted exome capture of 88 LUAD tumors from the NYU Center for Biospecimen Research and Development (XLSX 112 kb)
Supplementary Table 2
GEMM derived Keap1-mutant gene expression signature (z-score table) and top enriched gene sets from MSigDB curated and Oncosig collections. Genes with increasingly positive scores are over-expressed in Keap1-mutant samples whereas those with negative scores exhibit relatively lower expression in Keap1- mutant samples (magnitude denotes strength of a gene's expression correlation with the signature) (XLSX 528 kb)
Supplementary Table 3
Nrf2 core target gene set derived from the union of three published datasets and Nrf2-induced targets from individual datasets (XLSX 42 kb)
Supplementary Table 4
Human lung adenocarcinoma (TCGA) derived KEAP1- mutant gene expression signature (z-score table) and top enriched genes sets from MSigDB curated collections. Genes with increasingly positive scores are over-expressed in KEAP1-mutant samples whereas those with negative scores exhibit relatively lower expression in KEAP1-mutant samples (magnitude denotes strength of a gene's expression correlation with the signature) (XLSX 674 kb)
Supplementary Table 5
Univariate and Multivariate Cox regression analyses for overall survival in the TCGA lung adenocarcinoma patient cohort (XLSX 21 kb)
Supplementary Table 6
CRISPR/Cas9 Nrf2 transcriptional target screen containing sgRNA sequences, gene descriptions, and sgRNA scores (XLSX 17 kb)
Supplementary Table 7
Clinical and genetic features of PDX models (XLSX 22 kb)
Supplementary Table 8
Primer Sequences (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Romero, R., Sayin, V., Davidson, S. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23, 1362–1368 (2017). https://doi.org/10.1038/nm.4407
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4407
This article is cited by
-
Prediction of metabolites associated with somatic mutations in cancers by using genome-scale metabolic models and mutation data
Genome Biology (2024)
-
The pleiotropic functions of reactive oxygen species in cancer
Nature Cancer (2024)
-
Ferroptosis in cancer: From molecular mechanisms to therapeutic strategies
Signal Transduction and Targeted Therapy (2024)
-
AURKA emerges as a vulnerable target for KEAP1-deficient non-small cell lung cancer by activation of asparagine synthesis
Cell Death & Disease (2024)
-
Impact of LKB1 status on radiation outcome in patients with stage III non-small-cell lung cancer
Scientific Reports (2024)