Abstract
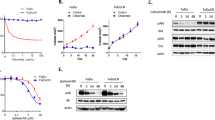
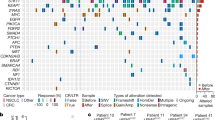
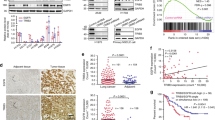
Targeting EGFR is a validated approach in the treatment of squamous-cell cancers (SCCs), although there are no established biomarkers for predicting response. We have identified a synonymous mutation in EGFR, c.2361G>A (encoding p.Gln787Gln), in two patients with head and neck SCC (HNSCC) who were exceptional responders to gefitinib, and we showed in patient-derived cultures that the A/A genotype was associated with greater sensitivity to tyrosine kinase inhibitors (TKIs) as compared to the G/A and G/G genotypes. Remarkably, single-copy G>A nucleotide editing in isogenic models conferred a 70-fold increase in sensitivity due to decreased stability of the EGFR-AS1 long noncoding RNA (lncRNA). In the appropriate context, sensitivity could be recapitulated through EGFR-AS1 knockdown in vitro and in vivo, whereas overexpression was sufficient to induce resistance to TKIs. Reduced EGFR-AS1 levels shifted splicing toward EGFR isoform D, leading to ligand-mediated pathway activation. In co-clinical trials involving patients and patient-derived xenograft (PDX) models, tumor shrinkage was most pronounced in the context of the A/A genotype for EGFR-Q787Q, low expression of EGFR-AS1 and high expression of EGFR isoform D. Our study reveals how a 'silent' mutation influences the levels of a lncRNA, resulting in noncanonical EGFR addiction, and delineates a new predictive biomarker suite for response to EGFR TKIs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cripps, C., Winquist, E., Devries, M.C., Stys-Norman, D. & Gilbert, R. Epidermal growth factor receptor targeted therapy in stages III and IV head and neck cancer. Curr. Oncol. 17, 37–48 (2010).
Montero, P.H. et al. Changing trends in smoking and alcohol consumption in patients with oral cancer treated at Memorial Sloan-Kettering Cancer Center from 1985 to 2009. Arch. Otolaryngol. Head Neck Surg. 138, 817–822 (2012).
Krishna Rao, S.V., Mejia, G., Roberts-Thomson, K. & Logan, R. Epidemiology of oral cancer in Asia in the past decade—an update (2000–2012). Asian Pac. J. Cancer Prev. 14, 5567–5577 (2013).
Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 45, 309–316 (2009).
Ng, J.H., Iyer, N.G., Tan, M.H. & Edgren, G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck 39, 297–304 (2017).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015).
Sacco, A.G. & Cohen, E.E. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J. Clin. Oncol. 33, 3305–3313 (2015).
Vermorken, J.B. et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 359, 1116–1127 (2008).
Echarri, M.J., Lopez-Martin, A. & Hitt, R. Targeted therapy in locally advanced and recurrent/metastatic head and neck squamous cell carcinoma (LA-R/M HNSCC). Cancers (Basel) 8, E27 (2016).
Licitra, L. et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann. Oncol. 22, 1078–1087 (2011).
Stewart, J.S. et al. Phase III study of gefitinib compared with intravenous methotrexate for recurrent squamous cell carcinoma of the head and neck. J. Clin. Oncol. 27, 1864–1871 (2009).
Kirby, A.M. et al. Gefitinib (ZD1839, Iressa) as palliative treatment in recurrent or metastatic head and neck cancer. Br. J. Cancer 94, 631–636 (2006).
Cohen, E.E. et al. Phase II trial of gefitinib 250 mg daily in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Clin. Cancer Res. 11, 8418–8424 (2005).
Thomas, F. et al. Pilot study of neoadjuvant treatment with erlotinib in nonmetastatic head and neck squamous cell carcinoma. Clin. Cancer Res. 13, 7086–7092 (2007).
Soulieres, D. et al. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J. Clin. Oncol. 22, 77–85 (2004).
Tan, D.S. et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer J. 15, 406–420 (2009).
Vettore, A.L. et al. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med. 7, 98 (2015).
Tan, D.S. et al. Tongue carcinoma infrequently harbor common actionable genetic alterations. BMC Cancer 14, 679 (2014).
Cohen, E.E. et al. Factors associated with clinical benefit from epidermal growth factor receptor inhibitors in recurrent and metastatic squamous cell carcinoma of the head and neck. Oral Oncol. 45, e155–e160 (2009).
Tan, E.H. et al. Gefitinib, cisplatin, and concurrent radiotherapy for locally advanced head and neck cancer: EGFR FISH, protein expression, and mutational status are not predictive biomarkers. Ann. Oncol. 23, 1010–1016 (2012).
Taguchi, T., Tsukuda, M., Imagawa-Ishiguro, Y., Kato, Y. & Sano, D. Involvement of EGFR in the response of squamous cell carcinoma of the head and neck cell lines to gefitinib. Oncol. Rep. 19, 65–71 (2008).
Leong, H.S. et al. Targeting cancer stem cell plasticity through modulation of epidermal growth factor and insulin-like growth factor receptor signaling in head and neck squamous cell cancer. Stem Cells Transl. Med. 3, 1055–1065 (2014).
Guillaudeau, A. et al. Adult diffuse gliomas produce mRNA transcripts encoding EGFR isoforms lacking a tyrosine kinase domain. Int. J. Oncol. 40, 1142–1152 (2012).
Adamczyk, K.A. et al. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sci. 89, 304–312 (2011).
Chia, S. et al. Phenotype-driven precision oncology—guiding clinical decisions one patient-at-a-time. Nat. Commun. (in the press).
Iyer, M.K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015).
Schmitt, A.M. & Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016).
Brown, J.A. et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640 (2014).
Qi, H.L. et al. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol. 37, 1079–1089 (2016).
Blythe, A.J., Fox, A.H. & Bond, C.S. The ins and outs of lncRNA structure: how, why and what comes next? Biochim. Biophys. Acta 1859, 46–58 (2016).
Ji, Z., Song, R., Regev, A. & Struhl, K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890 (2015).
Munroe, S.H. & Lazar, M.A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 266, 22083–22086 (1991).
Halle, C. et al. Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node–negative cervical cancer. Clin. Cancer Res. 17, 5501–5512 (2011).
Lococo, F. et al. Preliminary evidence on the diagnostic and molecular role of circulating soluble EGFR in non–small cell lung cancer. Int. J. Mol. Sci. 16, 19612–19630 (2015).
Romero-Ventosa, E.Y. et al. Pretreatment levels of the serum biomarkers CEA, CYFRA 21-1, SCC and the soluble EGFR and its ligands EGF, TGF-α, HB-EGF in the prediction of outcome in erlotinib treated non-small-cell lung cancer patients. Springerplus 4, 171 (2015).
Albitar, L. et al. EGFR isoforms and gene regulation in human endometrial cancer cells. Mol. Cancer 9, 166 (2010).
Zhou, T., Kim, Y. & MacLeod, A.R. Targeting long noncoding RNA with antisense oligonucleotide technology as cancer therapeutics. Methods Mol. Biol. 1402, 199–213 (2016).
Golan, T. et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 6, 24560–24570 (2015).
Tan, D.S., Mok, T.S. & Rebbeck, T.R. Cancer genomics: diversity and disparity across ethnicity and geography. J. Clin. Oncol. 34, 91–101 (2016).
Zhao, Y. et al. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene 27, 1–8 (2008).
Brumbaugh, C.D., Kim, H.J., Giovacchini, M. & Pourmand, N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinformatics 12, 479 (2011).
Garrison, P., Yue, S., Hanson, J., Baron, J. & Lui, J.C. Spatial regulation of bone morphogenetic proteins (BMPs) in postnatal articular and growth plate cartilage. PLoS One 12, e0176752 (2017).
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Acknowledgements
We would like to extend our gratitude to all patients and families involved in this study. We would also like to thank C. Chuan Young Ng of the National Cancer Centre Singapore Genomics Service for technical support with the NanoString assays. D.S.W.T. and N.G.I. are both supported by National Medical Research Council (NMRC) (Singapore) clinician–scientist awards (D.S.W.T.: NMRC/CSA/007/2016; N.G.I.: NMRC/CSA/042/2012, NMRC/CSA-INV/011/2016). Further support for this project was obtained from NMRC (grant no. NMRC/1304/2011, NMRC/CIRG/1434/2015), National Cancer Centre Research Funds and a Singhealth Foundation Grant (SHF/FG496P/2012).
Author information
Authors and Affiliations
Contributions
D.S.W.T. and N.G.I. conceived and designed the study. F.T.C., H.S.L., S.Y.T., D.P.L., X.L.K., X.Z., G.M.S. and G.S.T. performed experiments, with additional technical guidance and expertise from B.T.C., T.K.H.L., P.S., B.C. and R.D. M.M.C. and A.J.S. performed computational analysis. D.S.W.T., W.T.L., E.H.T. and M.K.A. conducted the co-clinical trials through the IMPACT protocol. F.T.C., H.S.L., S.Y.T. and N.G.I. performed statistical analyses. D.S.W.T. and N.G.I. wrote the manuscript, with extensive input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Figures 1–4 & Supplementary Tables 1–4 (PDF 21526 kb)
Supplementary Data
Supplementary Data (PDF 16045 kb)
Rights and permissions
About this article
Cite this article
Tan, D., Chong, F., Leong, H. et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med 23, 1167–1175 (2017). https://doi.org/10.1038/nm.4401
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4401
This article is cited by
-
Identification of m6A/m5C-related lncRNA signature for prediction of prognosis and immunotherapy efficacy in esophageal squamous cell carcinoma
Scientific Reports (2024)
-
Super enhancer loci of EGFR regulate EGFR variant 8 through enhancer RNA and strongly associate with survival in HNSCCs
Molecular Genetics and Genomics (2024)
-
LncRNA LINC01123 promotes malignancy of ovarian cancer by targeting hsa-miR-516b-5p/VEGFA
Genes & Genomics (2024)
-
A new marker constructed from immune-related lncRNA pairs can be used to predict clinical treatment effects and prognosis: in-depth exploration of underlying mechanisms in HNSCC
World Journal of Surgical Oncology (2023)
-
TUBA1C: a new potential target of LncRNA EGFR-AS1 promotes gastric cancer progression
BMC Cancer (2023)