Abstract
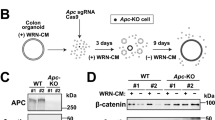
With the goal of modeling human disease of the large intestine, we sought to develop an effective protocol for deriving colonic organoids (COs) from differentiated human embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs). Extensive gene and immunohistochemical profiling confirmed that the derived COs represent colon rather than small intestine, containing stem cells, transit-amplifying cells, and the expected spectrum of differentiated cells, including goblet and endocrine cells. We applied this strategy to iPSCs derived from patients with familial adenomatous polyposis (FAP-iPSCs) harboring germline mutations in the WNT-signaling-pathway-regulator gene encoding APC, and we generated COs that exhibit enhanced WNT activity and increased epithelial cell proliferation, which we used as a platform for drug testing. Two potential compounds, XAV939 and rapamycin, decreased proliferation in FAP-COs, but also affected cell proliferation in wild-type COs, which thus limits their therapeutic application. By contrast, we found that geneticin, a ribosome-binding antibiotic with translational 'read-through' activity, efficiently targeted abnormal WNT activity and restored normal proliferation specifically in APC-mutant FAP-COs. These studies provide an efficient strategy for deriving human COs, which can be used in disease modeling and drug discovery for colorectal disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
Gene Expression Omnibus
Change history
13 March 2018
In the version of this article initially published, there were several instances of duplication of confocal microscopy images presented in Figures 1e, 3i and 4c,h,j. These errors occurred during the final preparation of the figures, when representative images were selected to illustrate the results obtained from multiple independent experiments. To ensure that these errors did not affect the paper’s conclusions, the authors verified all raw data used to quantify the results. The results presented in the original Figures 3j and 4d were not affected, but the quantification in Figure 4i was. This quantification was performed again, and the results supported the original conclusions of the paper. In addition, the western blot shown in Figure 3d came from a different replicate experiment than the one represented in Supplementary Figure 9g,h (which shows the raw data). The original blot has been replaced with the correct blot that matches the raw data originally provided. The conclusions of this experiment remain unchanged. All affected images have been now corrected in the HTML and PDF versions of the article.
References
Spence, J.R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
McCracken, K.W. et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014).
Lancaster, M.A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Dye, B.R. et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4, e05098 (2015).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Sherwood, R.I., Maehr, R., Mazzoni, E.O. & Melton, D.A. Wnt signaling specifies and patterns intestinal endoderm. Mech. Dev. 128, 387–400 (2011).
Pitera, J.E., Smith, V.V., Thorogood, P. & Milla, P.J. Coordinated expression of 3′ hox genes during murine embryonal gut development: an enteric Hox code. Gastroenterology 117, 1339–1351 (1999).
Bekku, S. et al. Carbonic anhydrase I and II as a differentiation marker of human and rat colonic enterocytes. Res. Exp. Med. (Berl.) 198, 175–185 (1998).
The Human Protein Atlas. CA4 (The Human Protein Atlas, n.d.); available at http://www.proteinatlas.org/ENSG00000167434–CA4/tissue/colon.
Batlle, E. et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111, 251–263 (2002).
Bretscher, A. & Weber, K. Villin: the major microfilament-associated protein of the intestinal microvillus. Proc. Natl. Acad. Sci. USA 76, 2321–2325 (1979).
Allen, A., Hutton, D.A. & Pearson, J.P. The MUC2 gene product: a human intestinal mucin. Int. J. Biochem. Cell Biol. 30, 797–801 (1998).
O'Connor, D.T., Burton, D. & Deftos, L.J. Chromogranin A: immunohistology reveals its universal occurrence in normal polypeptide hormone producing endocrine glands. Life Sci. 33, 1657–1663 (1983).
Roth, K.A., Kim, S. & Gordon, J.I. Immunocytochemical studies suggest two pathways for enteroendocrine cell differentiation in the colon. Am. J. Physiol. 263, G174–G180 (1992).
Legay, C., Faudon, M., Héry, F. & Ternaux, J.P. 5-HT metabolism in the intestinal wall of the rat-I. The mucosa. Neurochem. Int. 5, 721–727 (1983).
Yan, K.S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466–471 (2012).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011).
Jenny, M. et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21, 6338–6347 (2002).
Moll, R., Schiller, D.L. & Franke, W.W. Identification of protein IT of the intestinal cytoskeleton as a novel type I cytokeratin with unusual properties and expression patterns. J. Cell Biol. 111, 567–580 (1990).
Green, M.D. et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 29, 267–272 (2011).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Beuling, E. et al. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev. Biol. 322, 179–189 (2008).
Forster, R. et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports 2, 838–852 (2014).
Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838 (2016).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Sasikumar, R., Rejitha, J.R., Binumon, P.K. & Manoj, M. Role of heterozygous APC mutation in niche succession and initiation of colorectal cancer—a computational study. PLoS One 6, e22720 (2011).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Taketo, M.M. Apc gene knockout mice as a model for familial adenomatous polyposis. Prog. Exp. Tumor Res. 35, 109–119 (1999).
Yu, B., Lane, M.E., Pestell, R.G., Albanese, C. & Wadler, S. Downregulation of cyclin D1 alters cdk 4- and cdk 2-specific phosphorylation of retinoblastoma protein. Mol. Cell Biol. Res. Commun. 3, 352–359 (2000).
Voloshanenko, O. et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat. Commun. 4, 2610 (2013).
Durno, C.A. Colonic polyps in children and adolescents. Can. J. Gastroenterol. 21, 233–239 (2007).
Faller, W.J. et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500 (2015).
Huang, S.M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Ignatenko, N.A. et al. Combination chemoprevention of intestinal carcinogenesis in a murine model of familial adenomatous polyposis. Nutr. Cancer 60 (Suppl. 1), 30–35 (2008).
Floquet, C., Rousset, J.P. & Bidou, L. Readthrough of premature termination codons in the adenomatous polyposis coli gene restores its biological activity in human cancer cells. PLoS One 6, e24125 (2011).
Zilberberg, A., Lahav, L. & Rosin-Arbesfeld, R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 59, 496–507 (2010).
Dow, L.E. et al. Apc restoration promotes cellular differentiation and establishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015).
Choy, J.Y. et al. A resource of ribosomal RNA-depleted RNA-seq data from different normal adult and fetal human tissues. Sci. Data 2, 150063 (2015).
Borras, E. et al. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev. Res. (Phila.) 9, 417–427 (2016).
Kanth, P. et al. Gene signature in sessile serrated polyps identifies colon cancer subtype. Cancer Prev. Res. (Phila.) 9, 456–465 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Greene, A.E., Charney, J., Nichols, W.W. & Coriell, L.L. Species identity of insect cell lines. In Vitro 7, 313–322 (1972).
Acknowledgements
S.C. is funded by The New York Stem Cell Foundation (R-103) and NIH/NIDDK (1 DP2 DK098093-01, DP3DK111907-01). This study was supported in part by a shared facility contract from the New York Department of Health (NYSTEM, C029156; to T.E. and S.C.); a Tri-institutional Stem Cell Initiative grant (2013-001; to F. M. and S.C.); and a pilot grant from the Center for Advanced Digestion Care (CADC) at Weill Cornell Medical College (to S.C. and S.L.). S.C. is New York Stem Cell Foundation-Robertson Investigator. This work was also supported by grants R03CA176788 (US National Institutes of Health/National Cancer Institute), the MD Anderson Cancer Center Institutional Research Grant (IRG) Program (E.V.), and a gift from the Feinberg Family to E.V.; Cancer Prevention Educational Award (R25T CA057730, US National Institutes of Health/National Cancer Institute (to K.C.); Arnold O. Beckman postdoctoral fellowship to H.J.C. We thank H.E. Varmus (Weill Cornell Medical College) for his support and J. Jin at Weill Cornell Medical College for kindly providing human aortic smooth muscle cells. We are grateful for the technical support and advice provided by H.S. Ralph in the Cell Screening Core Facility, J. McCormick in the Flow Cytometry Facility and L. Cohen-Gould in the Electron Microscopy Facility at Weill Cornell Medical College.
Author information
Authors and Affiliations
Contributions
S.C., T.E., S.L., and F.R.M. designed the project; M.C., S.-Y.T., S.A., T.S., N.H.P.H.J.C., M.W., and M.G. performed experiments; E.V., K.C., T. Z., and J.Z.X. performed the bioinformatics analysis; M.C., E.V., K.C., and S.C. analyzed data; and M.C., E.V., F.R.M., S.L., T.E., and S.C. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–3 (PDF 42809 kb)
Rights and permissions
About this article
Cite this article
Crespo, M., Vilar, E., Tsai, SY. et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med 23, 878–884 (2017). https://doi.org/10.1038/nm.4355
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4355
This article is cited by
-
Integration of pan-omics technologies and three-dimensional in vitro tumor models: an approach toward drug discovery and precision medicine
Molecular Cancer (2024)
-
The role of organoids in cancer research
Experimental Hematology & Oncology (2023)
-
Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy
Molecular Cancer (2023)
-
Mechanically enhanced biogenesis of gut spheroids with instability-driven morphomechanics
Nature Communications (2023)
-
Functional precision oncology using patient-derived assays: bridging genotype and phenotype
Nature Reviews Clinical Oncology (2023)