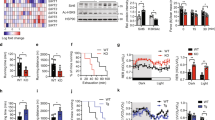
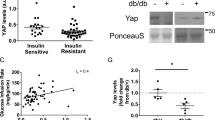
Abstract
Type 2 diabetes and insulin resistance are associated with reduced glucose utilization in the muscle and poor exercise performance. Here we find that depletion of the epigenome modifier histone deacetylase 3 (HDAC3) specifically in skeletal muscle causes severe systemic insulin resistance in mice but markedly enhances endurance and resistance to muscle fatigue, despite reducing muscle force. This seemingly paradoxical phenotype is due to lower glucose utilization and greater lipid oxidation in HDAC3-depleted muscles, a fuel switch caused by the activation of anaplerotic reactions driven by AMP deaminase 3 (Ampd3) and catabolism of branched-chain amino acids. These findings highlight the pivotal role of amino acid catabolism in muscle fatigue and type 2 diabetes pathogenesis. Further, as genome occupancy of HDAC3 in skeletal muscle is controlled by the circadian clock, these results delineate an epigenomic regulatory mechanism through which the circadian clock governs skeletal muscle bioenergetics. These findings suggest that physical exercise at certain times of the day or pharmacological targeting of HDAC3 could potentially be harnessed to alter systemic fuel metabolism and exercise performance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
DeFronzo, R.A. & Tripathy, D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 32 (Suppl. 2) S157–S163 (2009).
Samuel, V.T. & Shulman, G.I. Mechanisms for insulin resistance: common threads and missing links. Cell 148, 852–871 (2012).
Dubé, J.J. et al. Exercise-induced alterations in intramyocellular lipids and insulin resistance: the athlete's paradox revisited. Am. J. Physiol. Endocrinol. Metab. 294, E882–E888 (2008).
Brüning, J.C. et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569 (1998).
Kim, J.K. et al. Redistribution of substrates to adipose tissue promotes obesity in mice with selective insulin resistance in muscle. J. Clin. Invest. 105, 1791–1797 (2000).
Holloszy, J.O. “Deficiency” of mitochondria in muscle does not cause insulin resistance. Diabetes 62, 1036–1040 (2013).
Lowell, B.B. & Shulman, G.I. Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 (2005).
Szendroedi, J., Phielix, E. & Roden, M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 92–103 (2011).
Muoio, D.M. & Neufer, P.D. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 15, 595–605 (2012).
Randle, P.J., Garland, P.B., Hales, C.N. & Newsholme, E.A. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet 1, 785–789 (1963).
Hue, L. & Taegtmeyer, H. The Randle cycle revisited: a new head for an old hat. Am. J. Physiol. Endocrinol. Metab. 297, E578–E591 (2009).
Kelley, D.E. & Mandarino, L.J. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes 49, 677–683 (2000).
Muoio, D.M. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell 159, 1253–1262 (2014).
Zhang, R., Lahens, N.F., Ballance, H.I., Hughes, M.E. & Hogenesch, J.B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA 111, 16219–16224 (2014).
Kornmann, B., Schaad, O., Bujard, H., Takahashi, J.S. & Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007).
Hatori, M. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012).
Bass, J. & Takahashi, J.S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010).
Yin, L. & Lazar, M.A. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. 19, 1452–1459 (2005).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Sun, Z. et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942 (2012).
Sun, Z., Feng, D., Everett, L.J., Bugge, A. & Lazar, M.A. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harb. Symp. Quant. Biol. 76, 49–55 (2011).
Harfmann, B.D., Schroder, E.A. & Esser, K.A. Circadian rhythms, the molecular clock, and skeletal muscle. J. Biol. Rhythms 30, 84–94 (2015).
Mayeuf-Louchart, A., Staels, B. & Duez, H. Skeletal muscle functions around the clock. Diabetes Obes. Metab. 17 (Suppl. 1), 39–46 (2015).
Kang, S., Tsai, L.T.-Y. & Rosen, E.D. Nuclear Mechanisms of Insulin Resistance. Trends Cell Biol. 26, 341–351 (2016).
Bottomley, M.J. et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 283, 26694–26704 (2008).
Fischle, W. et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 (2002).
Haberland, M., Montgomery, R.L. & Olson, E.N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 (2009).
Bothe, G.W., Haspel, J.A., Smith, C.L., Wiener, H.H. & Burden, S.J. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis 26, 165–166 (2000).
Loro, E. et al. IL-15Rα is a determinant of muscle fuel utilization, and its loss protects against obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 309, R835–R844 (2015).
Zierath, J.R. & Hawley, J.A. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2, e348 (2004).
Schiaffino, S. & Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 (2011).
Nedachi, T., Fujita, H. & Kanzaki, M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 295, E1191–E1204 (2008).
Fang, B. et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014).
Lin, J. et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 (2002).
Calvo, J.A. et al. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J. Appl. Physiol. 104, 1304–1312 (2008).
Choi, C.S. et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc. Natl. Acad. Sci. USA 105, 19926–19931 (2008).
Wong, K.E. et al. Muscle-Specific Overexpression of PGC-1α Does Not Augment Metabolic Improvements in Response to Exercise and Caloric Restriction. Diabetes 64, 1532–1543 (2015).
Finck, B.N. et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 1, 133–144 (2005).
Pagel-Langenickel, I., Bao, J., Pang, L. & Sack, M.N. The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr. Rev. 31, 25–51 (2010).
Zhang, Y. et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015).
Woldt, E. et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013).
Yamamoto, H. et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 147, 827–839 (2011).
Lau, P., Fitzsimmons, R.L., Pearen, M.A., Watt, M.J. & Muscat, G.E.O. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia 54, 1169–1180 (2011).
Samant, S.A., Pillai, V.B., Sundaresan, N.R., Shroff, S.G. & Gupta, M.P. Histone Deacetylase 3 (HDAC3)-dependent Reversible Lysine Acetylation of Cardiac Myosin Heavy Chain Isoforms Modulates Their Enzymatic and Motor Activity. J. Biol. Chem. 290, 15559–15569 (2015).
Wagenmakers, A.J. Muscle amino acid metabolism at rest and during exercise: role in human physiology and metabolism. Exerc. Sport Sci. Rev. 26, 287–314 (1998).
Gibala, M.J., Young, M.E. & Taegtmeyer, H. Anaplerosis of the citric acid cycle: role in energy metabolism of heart and skeletal muscle. Acta Physiol. Scand. 168, 657–665 (2000).
Arinze, I.J. Facilitating understanding of the purine nucleotide cycle and the one-carbon pool: Part I: The purine nucleotide cycle. Biochem. Mol. Biol. Educ. 33, 165–168 (2005).
Bowtell, J.L., Marwood, S., Bruce, M., Constantin-Teodosiu, D. & Greenhaff, P.L. Tricarboxylic acid cycle intermediate pool size: functional importance for oxidative metabolism in exercising human skeletal muscle. Sports Med. 37, 1071–1088 (2007).
She, P. et al. Disruption of BCAA metabolism in mice impairs exercise metabolism and endurance. J. Appl. Physiol. 108, 941–949 (2010).
Ginevičienė, V. et al. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 15, 58 (2014).
Van den Berghe, G., Bontemps, F., Vincent, M.F. & Van den Bergh, F. The purine nucleotide cycle and its molecular defects. Prog. Neurobiol. 39, 547–561 (1992).
Operti, M.G., Vincent, M.F., Brucher, J.M. & van den Berghe, G. Enzymes of the purine nucleotide cycle in muscle of patients with exercise intolerance. Muscle Nerve 21, 401–403 (1998).
Templeman, N.M., Schutz, H., Garland, T. Jr. & McClelland, G.B. Do mice bred selectively for high locomotor activity have a greater reliance on lipids to power submaximal aerobic exercise? Am. J. Physiol. Regul. Integr. Comp. Physiol. 303, R101–R111 (2012).
Overmyer, K.A. et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 21, 468–478 (2015).
She, P. et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 6, 181–194 (2007).
Cheng, J. et al. AMPD1: a novel therapeutic target for reversing insulin resistance. BMC Endocr. Disord. 14, 96 (2014).
Jang, C. et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat. Med. 22, 421–426 (2016).
Hawley, J.A., Schabort, E.J., Noakes, T.D. & Dennis, S.C. Carbohydrate-loading and exercise performance. An update. Sports Med. 24, 73–81 (1997).
Phinney, S.D. Ketogenic diets and physical performance. Nutr. Metab. (Lond.) 1, 2 (2004).
Paoli, A., Bianco, A. & Grimaldi, K.A. The Ketogenic Diet and Sport: A Possible Marriage? Exerc. Sport Sci. Rev. 43, 153–162 (2015).
Cheng, R. et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 588, 3954–3958 (2014).
Lu, W. et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 (2010).
Melamud, E., Vastag, L. & Rabinowitz, J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 (2010).
Sun, Z. et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 (2013).
Acknowledgements
We thank S. Burden (New York University) for MLC-Cre mice, J. Hogenesch (University of Pennsylvania) for processed data from circaDB, M. Birnbaum and J. Baur (University of Pennsylvania) for helpful discussion, S. Guo (Texas A&M University) for help with the EPS procedure, P. Zhang (Baylor College of Medicine) for adenoviral plasmids, S. Hui and J. Park (Princeton University) for analysis of metabolomics data, V. Narkar (University of Texas Health Science Center) for the immunostaining protocol, M. Goncalves and C. Lanzillotta (University of Pennsylvania) for technical assistance. We thank the Penn Diabetes Center (DK19525) Functional Genomics Core for nucleotide sequencing, the Mouse Metabolic Phenotyping Core for clamp experiments, the Penn Muscle Institute Muscle Physiology Assessment Core for muscle contraction study, and the Princeton/Penn Regional Metabolomics Core for flux and lipid analysis. We thank the Baylor Diabetes Center (DK079638) Metabolism Core for Seahorse analysis and Vanderbilt MMPC (DK59637) for lipidomics analysis. The Rockefeller Proteomics Resource Center is supported by the Leona M. and Harry B. Helmsley Charitable Trust. This work was supported by NIH grants CA211437 (W.L.), DK043806 (M.A.L.) and DK099443 (Z.S.).
Author information
Authors and Affiliations
Contributions
Z.S. and M.A.L. conceived the study and designed experiments. S.H., W.Z., W.L., E.L., G.D., J.J., S.Z., Y.Z., D.F., Q.C., B.D.D. and Z.S. conducted experiments. S.H., W.Z., B.F., W.L., M.D., G.D., S.Z., B.D.D., H.M. and Z.S. analyzed the data. E.L., T.S.K., J.D.R., M.A.L. and Z.S. interpreted the data. M.A.L. and Z.S. acquired funding. Z.S. wrote the manuscript with input from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
Supplementary Figures 1–7 (PDF 2216 kb)
Supplementary Table 1
Raw data of metabolomics study. (XLSX 27 kb)
Supplementary Table 2
Differentially expressed genes from RNA-seq. (XLSX 44 kb)
Supplementary Table 3
Differentially expressed proteins from proteomics. (XLSX 475 kb)
Rights and permissions
About this article
Cite this article
Hong, S., Zhou, W., Fang, B. et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat Med 23, 223–234 (2017). https://doi.org/10.1038/nm.4245
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4245
This article is cited by
-
Role of AMP deaminase in diabetic cardiomyopathy
Molecular and Cellular Biochemistry (2024)
-
Epstein–Barr virus-acquired immunodeficiency in myalgic encephalomyelitis—Is it present in long COVID?
Journal of Translational Medicine (2023)
-
Daytime-restricted feeding enhances running endurance without prior exercise in mice
Nature Metabolism (2023)
-
The role of HDAC3 and its inhibitors in regulation of oxidative stress and chronic diseases
Cell Death Discovery (2023)
-
Epigenetics of type 2 diabetes mellitus and weight change — a tool for precision medicine?
Nature Reviews Endocrinology (2022)