Abstract
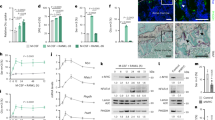
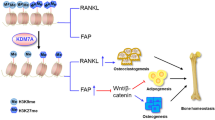
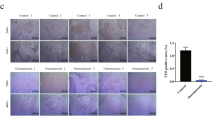
Metabolic reprogramming occurs in response to the cellular environment to mediate differentiation1,2,3, but the fundamental mechanisms linking metabolic processes to differentiation programs remain to be elucidated. During osteoclast differentiation, a shift toward more oxidative metabolic processes occurs3. In this study we identified the de novo DNA methyltransferase 3a (Dnmt3a) as a transcription factor that couples these metabolic changes to osteoclast differentiation. We also found that receptor activator of nuclear factor-κB ligand (RANKL), an essential cytokine for osteoclastogenesis4,5,6,7, induces this metabolic shift towards oxidative metabolism, which is accompanied by an increase in S-adenosylmethionine (SAM) production. We found that SAM-mediated DNA methylation by Dnmt3a regulates osteoclastogenesis via epigenetic repression of anti-osteoclastogenic genes. The importance of Dnmt3a in bone homeostasis was underscored by the observations that Dnmt3a-deficient osteoclast precursor cells do not differentiate efficiently into osteoclasts and that mice with an osteoclast-specific deficiency in Dnmt3a have elevated bone mass due to a smaller number of osteoclasts. Furthermore, inhibition of DNA methylation by theaflavin-3,3′-digallate abrogated bone loss in models of osteoporosis. Thus, this study reveals the role of epigenetic processes in the regulation of cellular metabolism and differentiation, which may provide the molecular basis for a new therapeutic strategy for a variety of bone disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Teperino, R., Schoonjans, K. & Auwerx, J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 12, 321–327 (2010).
Lu, C. & Thompson, C.B. Metabolic regulation of epigenetics. Cell Metab. 16, 9–17 (2012).
Ishii, K.A. et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat. Med. 15, 259–266 (2009).
Theill, L.E., Boyle, W.J. & Penninger, J.M. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu. Rev. Immunol. 20, 795–823 (2002).
Xing, L., Schwarz, E.M. & Boyce, B.F. Osteoclast precursors, RANKL/RANK, and immunology. Immunol. Rev. 208, 19–29 (2005).
Lorenzo, J., Horowitz, M. & Choi, Y. Osteoimmunology: interactions of the bone and immune system. Endocr. Rev. 29, 403–440 (2008).
Jones, D., Glimcher, L.H. & Aliprantis, A.O. Osteoimmunology at the nexus of arthritis, osteoporosis, cancer, and infection. J. Clin. Invest. 121, 2534–2542 (2011).
Karsenty, G. & Wagner, E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2, 389–406 (2002).
Yasui, T., Hirose, J., Aburatani, H. & Tanaka, S. Epigenetic regulation of osteoclast differentiation. Ann. NY Acad. Sci. 1240, 7–13 (2011).
Ross, F.P. & Teitelbaum, S.L. αvβ3 and macrophage colony-stimulating factor: partners in osteoclast biology. Immunol. Rev. 208, 88–105 (2005).
Hayashi, M. et al. Osteoprotection by semaphorin 3A. Nature 485, 69–74 (2012).
Nishikawa, K. et al. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. USA 107, 3117–3122 (2010).
Mund, C., Brueckner, B. & Lyko, F. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics 1, 7–13 (2006).
Rajavelu, A., Tulyasheva, Z., Jaiswal, R., Jeltsch, A. & Kuhnert, N. The inhibition of the mammalian DNA methyltransferase 3a (Dnmt3a) by dietary black tea and coffee polyphenols. BMC Biochem. 12, 16 (2011).
Okano, M., Bell, D.W., Haber, D.A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Nguyen, S., Meletis, K., Fu, D., Jhaveri, S. & Jaenisch, R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 236, 1663–1676 (2007).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Nakamura, T. et al. Estrogen prevents bone loss via estrogen receptor α and induction of Fas ligand in osteoclasts. Cell 130, 811–823 (2007).
Maeda, K. et al. Wnt5a–Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat. Med. 18, 405–412 (2012).
Dacquin, R., Starbuck, M., Schinke, T. & Karsenty, G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 224, 245–251 (2002).
Wu, H. et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448 (2010).
Rodan, G.A. & Martin, T.J. Therapeutic approaches to bone diseases. Science 289, 1508–1514 (2000).
Goldring, S.R. & Gravallese, E.M. Bisphosphonates: environmental protection for the joint? Arthritis Rheum. 50, 2044–2047 (2004).
Negishi-Koga, T. et al. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat. Med. 17, 1473–1480 (2011).
Shinohara, M. et al. Tyrosine kinases Btk and Tec regulate osteoclast differentiation by linking RANK and ITAM signals. Cell 132, 794–806 (2008).
Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 (2002).
de la Rica, L. et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 (2013).
Teo, B.H., Bobryshev, Y.V., Teh, B.K., Wong, S.H. & Lu, J. Complement C1q production by osteoclasts and its regulation of osteoclast development. Biochem. J. 447, 229–237 (2012).
Hikiji, H. et al. TDAG8 activation inhibits osteoclastic bone resorption. FASEB J. 28, 871–879 (2014).
Zhao, B. et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 (2009).
Zhang, Y. et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38, 4246–4253 (2010).
Paro, R. Chromatin regulation. Formatting genetic text. Nature 406, 579–580 (2000).
Fujikawa, M. & Yoshida, M. A sensitive, simple assay of mitochondrial ATP synthesis of cultured mammalian cells suitable for high-throughput analysis. Biochem. Biophys. Res. Commun. 401, 538–543 (2010).
Rodríguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339 (2011).
Selander, K., Lehenkari, P. & Vaananen, H.K. The effects of bisphosphonates on the resorption cycle of isolated osteoclasts. Calcif. Tissue Int. 55, 368–375 (1994).
Nishikawa, K. et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J. Clin. Invest. 120, 3455–3465 (2010).
Nishikawa, K., Iwamoto, Y. & Ishii, M. Development of an in vitro culture method for stepwise differentiation of mouse embryonic stem cells and induced pluripotent stem cells into mature osteoclasts. J. Bone Miner. Metab. 32, 331–336 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Weber, M. et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37, 853–862 (2005).
Xu, Y. et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151, 1200–1213 (2012).
Murrell, A., Heeson, S. & Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 (2004).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000).
Nishikawa, K. et al. Self-association of Gata1 enhances transcriptional activity in vivo in zebra fish embryos. Mol. Cell. Biol. 23, 8295–8305 (2003).
Acknowledgements
We thank K. Kaseda, M. Shirazaki, and Y. Maijima for technical assistance and J. Kikuta, S. Fujimori, and S. Simmons for helpful discussions. We also thank M. Okano, S. Takeda, and G. Karsenty; H. Wu, M. Takami, and K. Ozato for the gifts of Dnmt3a-deficient ESCs, Col1a1–Cre transgenic mice, the Dnmt3a expression vector, and Irf8−/− mice, respectively. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas from the Japan Society for the Promotion of Science (JSPS) (26116719; K.N.); Grants-in-Aid for Creative Scientific Research and Young Scientists (A) from the JSPS (26713010; K.N.); Grants-in-Aid for Scientific Research (A) from the JSPS (25253070; M.I.); the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT; M.I.); the Platform for Drug Discovery, Informatics, and Structural Life Science from the MEXT, Japan (T.T., S. Kawaguchi and M.Y.); and grants from the Astellas Foundation for Research on Metabolic Disorders (K.N.), the Ichiro Kanehara Foundation (K.N.), the Shimadzu Science Foundation (K.N.), the Takeda Science Foundation (K.N. and M.I.), and the International Human Frontier Science Program (RGY0077/2011 to M.I.).
Author information
Authors and Affiliations
Contributions
K.N. directed the project, conducted most of the experiments, and prepared the manuscript. Y.I. contributed to in vitro analyses. Y.K. supported the generation of osteoclast-specific Dnmt3a knockout mice. F.K. and M.Y. contributed to MBD-seq analyses. S. Kawaguchi and T.T. contributed to in silico analyses. T.N. and S. Kato generated CtskCre/+ mice. H.T. supported the GeneChip analysis. M.I. helped to direct the project and prepare the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 (PDF 9150 kb)
Supplementary Data Set 1
Supplementary Table 1 (XLS 47 kb)
Supplementary Data Set 2
Supplementary Table 2 (XLS 319 kb)
Rights and permissions
About this article
Cite this article
Nishikawa, K., Iwamoto, Y., Kobayashi, Y. et al. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine–producing metabolic pathway. Nat Med 21, 281–287 (2015). https://doi.org/10.1038/nm.3774
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3774
This article is cited by
-
The serine synthesis pathway drives osteoclast differentiation through epigenetic regulation of NFATc1 expression
Nature Metabolism (2024)
-
Novel Insights into Osteoclast Energy Metabolism
Current Osteoporosis Reports (2023)
-
Pivotal role for S-nitrosylation of DNA methyltransferase 3B in epigenetic regulation of tumorigenesis
Nature Communications (2023)
-
Osteoblast-derived vesicles induce a switch from bone-formation to bone-resorption in vivo
Nature Communications (2022)
-
TGFβ reprograms TNF stimulation of macrophages towards a non-canonical pathway driving inflammatory osteoclastogenesis
Nature Communications (2022)