Abstract
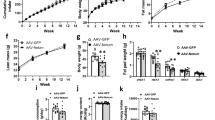
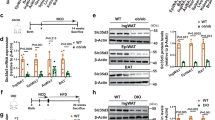
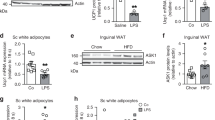
Brown fat activates uncoupled respiration in response to cold temperature and contributes to systemic metabolic homeostasis. To date, the metabolic action of brown fat has been primarily attributed to its role in fuel oxidation and uncoupling protein 1 (UCP1)-mediated thermogenesis. Whether brown fat engages other tissues through secreted factors remains largely unexplored. Here we show that neuregulin 4 (Nrg4), a member of the epidermal growth factor (EGF) family of extracellular ligands, is highly expressed in adipose tissues, enriched in brown fat and markedly increased during brown adipocyte differentiation. Adipose tissue Nrg4 expression was reduced in rodent and human obesity. Gain- and loss-of-function studies in mice demonstrated that Nrg4 protects against diet-induced insulin resistance and hepatic steatosis through attenuating hepatic lipogenic signaling. Mechanistically, Nrg4 activates ErbB3 and ErbB4 signaling in hepatocytes and negatively regulates de novo lipogenesis mediated by LXR and SREBP1c in a cell-autonomous manner. These results establish Nrg4 as a brown fat–enriched endocrine factor with therapeutic potential for the treatment of obesity-associated disorders, including type 2 diabetes and nonalcoholic fatty liver disease (NAFLD).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Kozak, L.P. & Harper, M.E. Mitochondrial uncoupling proteins in energy expenditure. Annu. Rev. Nutr. 20, 339–363 (2000).
Lowell, B.B. et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366, 740–742 (1993).
Yoneshiro, T. et al. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 123, 3404–3408 (2013).
Bartelt, A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011).
van der Lans, A.A. et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 123, 3395–3403 (2013).
Nedergaard, J., Bengtsson, T. & Cannon, B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293, E444–E452 (2007).
Cypess, A.M. et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009).
van Marken Lichtenbelt, W.D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Virtanen, K.A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009).
Enerbäck, S. Human brown adipose tissue. Cell Metab. 11, 248–252 (2010).
Nedergaard, J. & Cannon, B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 11, 268–272 (2010).
Petrovic, N. et al. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285, 7153–7164 (2010).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Cypess, A.M. et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639 (2013).
Jespersen, N.Z. et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 17, 798–805 (2013).
Seale, P. et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967 (2008).
Enerbäck, S. et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997).
Feldmann, H.M., Golozoubova, V., Cannon, B. & Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009).
Liu, X. et al. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. J. Clin. Invest. 111, 399–407 (2003).
Kadowaki, T. et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Invest. 116, 1784–1792 (2006).
Trujillo, M.E. & Scherer, P.E. Adipose tissue-derived factors: impact on health and disease. Endocr. Rev. 27, 762–778 (2006).
Waki, H. & Tontonoz, P. Endocrine functions of adipose tissue. Annu. Rev. Pathol. 2, 31–56 (2007).
Angelin, B., Larsson, T.E. & Rudling, M. Circulating fibroblast growth factors as metabolic regulators—a critical appraisal. Cell Metab. 16, 693–705 (2012).
Potthoff, M.J., Kliewer, S.A. & Mangelsdorf, D.J. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 26, 312–324 (2012).
Pedersen, B.K. & Febbraio, M.A. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 8, 457–465 (2012).
Kim, K.H. et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 19, 83–92 (2013).
Karsenty, G. & Ferron, M. The contribution of bone to whole-organism physiology. Nature 481, 314–320 (2012).
Schneider, M.R. & Wolf, E. The epidermal growth factor receptor ligands at a glance. J. Cell. Physiol. 218, 460–466 (2009).
Holbro, T. & Hynes, N.E. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 44, 195–217 (2004).
Bublil, E.M. & Yarden, Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr. Opin. Cell Biol. 19, 124–134 (2007).
Chen, Y. et al. SPD—a web-based secreted protein database. Nucleic Acids Res. 33, D169–D173 (2005).
Odiete, O., Hill, M.F. & Sawyer, D.B. Neuregulin in cardiovascular development and disease. Circ. Res. 111, 1376–1385 (2012).
Hayes, N.V., Newsam, R.J., Baines, A.J. & Gullick, W.J. Characterization of the cell membrane-associated products of the neuregulin 4 gene. Oncogene 27, 715–720 (2008).
Harari, D. et al. Neuregulin-4: a novel growth factor that acts through the ErbB-4 receptor tyrosine kinase. Oncogene 18, 2681–2689 (1999).
Müller, H., Dai, G. & Soares, M.J. Placental lactogen-I (PL-I) target tissues identified with an alkaline phosphatase-PL-I fusion protein. J. Histochem. Cytochem. 46, 737–743 (1998).
Horton, J.D., Goldstein, J.L. & Brown, M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Carver, R.S., Stevenson, M.C., Scheving, L.A. & Russell, W.E. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology 123, 2017–2027 (2002).
Olayioye, M.A., Beuvink, I., Horsch, K., Daly, J.M. & Hynes, N.E. ErbB receptor–induced activation of stat transcription factors is mediated by Src tyrosine kinases. J. Biol. Chem. 274, 17209–17218 (1999).
Jones, F.E., Welte, T., Fu, X.Y. & Stern, D.F. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 147, 77–88 (1999).
Repa, J.J. et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 (2000).
Schultz, J.R. et al. Role of LXRs in control of lipogenesis. Genes Dev. 14, 2831–2838 (2000).
Stöcklin, E., Wissler, M., Gouilleux, F. & Groner, B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383, 726–728 (1996).
Gregor, M.F. & Hotamisligil, G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445 (2011).
Odegaard, J.I. & Chawla, A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science 339, 172–177 (2013).
Osborn, O. & Olefsky, J.M. The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 18, 363–374 (2012).
Hotamisligil, G.S., Shargill, N.S. & Spiegelman, B.M. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science 259, 87–91 (1993).
Kohjima, M. et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int. J. Mol. Med. 21, 507–511 (2008).
Shimomura, I., Bashmakov, Y. & Horton, J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 274, 30028–30032 (1999).
Knebel, B. et al. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS ONE 7, e31812 (2012).
Shimano, H. et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99, 846–854 (1997).
Tang, J.J. et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13, 44–56 (2011).
Moon, Y.A. et al. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 15, 240–246 (2012).
Rosell, M. et al. Brown and white adipose tissues. Intrinsic differences in gene expression and response to cold exposure in mice. Am. J. Physiol. Endocrinol. Metab. 306, E945–E964 (2014).
Klein, J., Fasshauer, M., Klein, H.H., Benito, M. & Kahn, C.R. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. Bioessays 24, 382–388 (2002).
Lin, J. & Linzer, D.I. Induction of megakaryocyte differentiation by a novel pregnancy-specific hormone. J. Biol. Chem. 274, 21485–21489 (1999).
Leahy, D.J., Dann, C.E. III, Longo, P., Perman, B. & Ramyar, K.X. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr. Purif. 20, 500–506 (2000).
Lin, J. et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119, 121–135 (2004).
Li, S. et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 8, 105–117 (2008).
Acknowledgements
We thank P. Dempsey (University of Colorado) for ErbB4-Min6 cells, Y. Yarden (Weizmann Institute of Science) for ErbB expression plasmids, D. Leahy (Johns Hopkins University) for expression constructs for ErbB3 and ErbB4 extracellular domains, D. Threadgill (Texas A&M University) for Erbb3flox/flox mice and A. Saltiel for discussions. We are grateful to Q. Yu and L. Wang for technical support, the lab members for discussion, and the staff at the University of Michigan Transgenic Animal Model Core and the metabolic phenotyping core supported by Michigan Diabetes Research and Training Center (DK020572) and Nutrition Obesity Research Center (DK089503). This work was supported by the US National Institutes of Health (DK077086 and DK095151, J.D.L.; DK097608, X.S.) and a grant from Novo Nordisk. S.L. and G.-X.W. were supported by Scientist Development Grant and Predoctoral Fellowship, respectively, from the American Heart Association.
Author information
Authors and Affiliations
Contributions
J.D.L., G.-X.W. and X.-Y.Z. conceived the project and designed research. G.-X.W., X.-Y.Z., Z. Chen, Z. Cozacov, S.L. and Z.-X.M. performed metabolic and molecular studies; M.K., A.D. and M.B. performed human studies. D.Z., A.L.O. and X.S. performed lipid profile analysis. G.-X.W. and J.D.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
This work was partially supported by a research agreement with Novo Nordisk.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 6608 kb)
Rights and permissions
About this article
Cite this article
Wang, GX., Zhao, XY., Meng, ZX. et al. The brown fat–enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 20, 1436–1443 (2014). https://doi.org/10.1038/nm.3713
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3713
This article is cited by
-
Lipid metabolic reprogramming mediated by circulating Nrg4 alleviates metabolic dysfunction-associated steatotic liver disease during the early recovery phase after sleeve gastrectomy
BMC Medicine (2024)
-
Integrated traditional Chinese and Western medicine in the prevention and treatment of non-alcoholic fatty liver disease: future directions and strategies
Chinese Medicine (2024)
-
Association of non-high-density lipoprotein cholesterol trajectories with the development of non-alcoholic fatty liver disease: an epidemiological and genome-wide association study
Journal of Translational Medicine (2023)
-
Correlation of asprosin and Nrg-4 with type 2 diabetes Mellitus Complicated with Coronary Heart Disease and the Diagnostic Value
BMC Endocrine Disorders (2023)
-
Cold-activated brown fat-derived extracellular vesicle-miR-378a-3p stimulates hepatic gluconeogenesis in male mice
Nature Communications (2023)