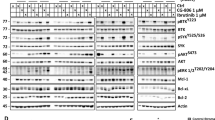
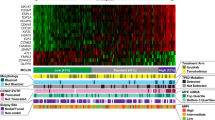
Abstract
Mantle cell lymphoma (MCL) is an aggressive malignancy that is characterized by poor prognosis1. Large-scale pharmacological profiling across more than 100 hematological cell line models identified a subset of MCL cell lines that are highly sensitive to the B cell receptor (BCR) signaling inhibitors ibrutinib and sotrastaurin. Sensitive MCL models exhibited chronic activation of the BCR-driven classical nuclear factor-κB (NF-κB) pathway, whereas insensitive cell lines displayed activation of the alternative NF-κB pathway. Transcriptome sequencing revealed genetic lesions in alternative NF-κB pathway signaling components in ibrutinib-insensitive cell lines, and sequencing of 165 samples from patients with MCL identified recurrent mutations in TRAF2 or BIRC3 in 15% of these individuals. Although they are associated with insensitivity to ibrutinib, lesions in the alternative NF-κB pathway conferred dependence on the protein kinase NIK (also called mitogen-activated protein 3 kinase 14 or MAP3K14) both in vitro and in vivo. Thus, NIK is a new therapeutic target for MCL treatment, particularly for lymphomas that are refractory to BCR pathway inhibitors. Our findings reveal a pattern of mutually exclusive activation of the BCR–NF-κB or NIK–NF-κB pathways in MCL and provide critical insights into patient stratification strategies for NF-κB pathway–targeted agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ghielmini, M. & Zucca, E. How I treat mantle cell lymphoma. Blood 114, 1469–1476 (2009).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Jabbour, E. & Kantarjian, H. Chronic myeloid leukemia: 2012 update on diagnosis, monitoring, and management. Am. J. Hematol. 87, 1037–1045 (2012).
Honigberg, L.A. et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA 107, 13075–13080 (2010).
Wagner, J. et al. Structure-activity relationship and pharmacokinetic studies of sotrastaurin (AEB071), a promising novel medicine for prevention of graft rejection and treatment of psoriasis. J. Med. Chem. 54, 6028–6039 (2011).
Wagner, J. et al. Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes. J. Med. Chem. 52, 6193–6196 (2009).
Naylor, T.L. et al. Protein kinase C inhibitor sotrastaurin selectively inhibits the growth of CD79 mutant diffuse large B-cell lymphomas. Cancer Res. 71, 2643–2653 (2011).
Davis, R.E. et al. Chronic active B-cell–receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 (2010).
Thome, M., Charton, J.E., Pelzer, C. & Hailfinger, S. Antigen receptor signaling to NF-κB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2, a003004 (2010).
Hailfinger, S. et al. Essential role of MALT1 protease activity in activated B cell–like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 106, 19946–19951 (2009).
Hailfinger, S. et al. Malt1-dependent RelB cleavage promotes canonical NF-κB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. USA 108, 14596–14601 (2011).
Lenz, G. et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 (2008).
Brockman, J.A. et al. Coupling of a signal response domain in IκBα to multiple pathways for NF-κ B activation. Mol. Cell Biol. 15, 2809–2818 (1995).
Chen, Z. et al. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 (1995).
Lam, L.T. et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-κB pathways in subtypes of diffuse large B-cell lymphoma. Blood 111, 3701–3713 (2008).
Lam, L.T. et al. Small molecule inhibitors of IκB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin. Cancer Res. 11, 28–40 (2005).
Annunziata, C.M. et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Zarnegar, B., Yamazaki, S., He, J.Q. & Cheng, G. Control of canonical NF-κB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. USA 105, 3503–3508 (2008).
Vallabhapurapu, S. et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat. Immunol. 9, 1364–1370 (2008).
Zarnegar, B.J. et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9, 1371–1378 (2008).
Ling, L., Cao, Z. & Goeddel, D.V. NF-κB–inducing kinase activates IKK-α by phosphorylation of Ser-176. Proc. Natl. Acad. Sci. USA 95, 3792–3797 (1998).
Senftleben, U. et al. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293, 1495–1499 (2001).
Claudio, E., Brown, K., Park, S., Wang, H. & Siebenlist, U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat. Immunol. 3, 958–965 (2002).
Dejardin, E. et al. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17, 525–535 (2002).
Demchenko, Y.N. et al. Classical and/or alternative NF-κB pathway activation in multiple myeloma. Blood 115, 3541–3552 (2010).
Demchenko, Y.N. & Kuehl, W.M. A critical role for the NFkB pathway in multiple myeloma. Oncotarget 1, 59–68 (2010).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Wang, M.L. et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 369, 507–516 (2013).
Malinin, N.L., Boldin, M.P., Kovalenko, A.V. & Wallach, D. MAP3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature 385, 540–544 (1997).
Ngo, V.N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006).
Nogai, H. et al. IκB-ζ controls the constitutive NF-κB target gene network and survival of ABC DLBCL. Blood 122, 2242–2250 (2013).
Pfeifer, M. et al. PTEN loss defines a PI3K/AKT pathway–dependent germinal center subtype of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 110, 12420–12425 (2013).
Ranuncolo, S.M., Pittaluga, S., Evbuomwan, M.O., Jaffe, E.S. & Lewis, B.A. Hodgkin lymphoma requires stabilized NIK and constitutive RelB expression for survival. Blood 120, 3756–3763 (2012).
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009).
Wee, S. et al. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. USA 105, 13057–13062 (2008).
Liao, G., Zhang, M., Harhaj, E.W. & Sun, S.C. Regulation of the NF-κB–inducing kinase by tumor necrosis factor receptor-associated factor 3–induced degradation. J. Biol. Chem. 279, 26243–26250 (2004).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Forbes, S.A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D.C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Mills, R.E. et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 21, 830–839 (2011).
Abecasis, G.R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Thorvaldsdóttir, H., Robinson, J.T. & Mesirov, J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Acknowledgements
We thank T. Haferlach and W. Kern from the Munich Leukemia Laboratory (MLL) for providing MCL samples. We thank N. Clipstone (Loyola University) for the pMSCV-IRES-GFP vector. We also thank M. Wartmann, M. Stump, T. Hood and S. Zhu for sharing technical advice. We are grateful to D. Weinstock, R. Pagliarini, D. Porter, S. Sharma, N. Wilson and N. Keen for their critical reading of this manuscript. All authors except M.F., R.K., F.C.C., B.M., K.D., B.D., C.S., A.T., M.H., R.D.G. and G.L. are employees of Novartis Institutes for Biomedical Research. This work was supported by research grants to G.L. from the Else Kröner-Fresenius-Stiftung, the German Research Foundation and the Deutsche Krebshilfe. Additionally, this work was supported by a New Frontiers in Cancer Terry Fox program project grant (19001) to R.D.G. and a Career Investigator award by the Michael Smith Foundation for Health Research to C.S. R.K. is a recipient of fellowships from the Canadian Institutes of Health Research, the Michael Smith Foundation for Health Research and the University of British Columbia (Four Year Fellowship).
Author information
Authors and Affiliations
Contributions
R. Rahal, M.F., W.R.S., G.L. and F.S. designed experiments and wrote the manuscript. R. Rahal, M.F., K.D. and R. Romero conducted in vitro experiments. H.B. and V.G.C. designed and conducted in vivo experiments. C.F., A.K., R. Rahal and F.S. designed the pharmacological screen. A.F., E.P. and R. Romero conducted the experiments. D. Ruddy and D. Rakiec performed RNA-seq experiments with input from A.D., J.M. and M.P.M. J.M.K. developed the methods to analyze RNA-seq data. T.N., S. Kovats and S. Kim contributed to the initial finding of the sensitivity of MCL to ibrutinib/STN. R.K., F.C.C., B.M. and R.D.G. designed, performed and analyzed targeted sequencing studies on samples from patients with MCL. C.S., B.D., A.T. and M.H. provided and characterized patient samples.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 8633 kb)
Supplementary Table 1
1A: Pharmacological profiling of 119 lymphoma and leukemia cell lines 1B: Pharmacological profiling of MCL cell lines (XLS 281 kb)
Supplementary Table 2
Gene modulated by AFN700 treatment (XLSX 6357 kb)
Supplementary Table 3
Nucleotide variants identified by RNA-seq , related to Figure 3 (XLS 5052 kb)
Supplementary Table 4
RPKM values derived from RNA-seq, related to Figure 3 (XLS 8224 kb)
Supplementary Table 5
Maver-1 copy number analysis by SNP 6.0 arrays (XLS 4946 kb)
Supplementary Table 6
Genes downregulated upon knockdown of NIK in both Z-138 and Maver-1 cells (XLS 39 kb)
Supplementary Table 7
Mutation calls in BIRC2, BIRC3, TRAF2, TRAF3 and MAP3K14 for 165 primary patient samples and 3 cell lines (XLS 46 kb)
Supplementary Table 8
shRNA sequences used in the study (XLS 33 kb)
Supplementary Table 9
Primary antibodies used in the study (XLS 32 kb)
Supplementary Table 10
Quantitative PCR probes used in the study (XLS 24 kb)
Supplementary Table 11
PCR primers used in the study (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Rahal, R., Frick, M., Romero, R. et al. Pharmacological and genomic profiling identifies NF-κB–targeted treatment strategies for mantle cell lymphoma. Nat Med 20, 87–92 (2014). https://doi.org/10.1038/nm.3435
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3435
This article is cited by
-
Beyond Bruton’s tyrosine kinase inhibitors in mantle cell lymphoma: bispecific antibodies, antibody–drug conjugates, CAR T-cells, and novel agents
Journal of Hematology & Oncology (2023)
-
MALT1-dependent cleavage of CYLD promotes NF-κB signaling and growth of aggressive B-cell receptor-dependent lymphomas
Blood Cancer Journal (2023)
-
TRAF2/3 deficient B cells resist DNA damage-induced apoptosis via NF-κB2/XIAP/cIAP2 axis and IAP antagonist sensitizes mutant lymphomas to chemotherapeutic drugs
Cell Death & Disease (2023)
-
The many faces of nodal and splenic marginal zone lymphomas. A report of the 2022 EA4HP/SH lymphoma workshop
Virchows Archiv (2023)
-
BTK inhibitors in the treatment of hematological malignancies and inflammatory diseases: mechanisms and clinical studies
Journal of Hematology & Oncology (2022)