Abstract
Pandemic influenza viruses often cause severe disease in middle-aged adults without preexisting comorbidities. The mechanism of illness associated with severe disease in this age group is not well understood1,2,3,4,5,6,7,8,9,10. Here we find preexisting serum antibodies that cross-react with, but do not protect against, 2009 H1N1 influenza virus in middle-aged adults. Nonprotective antibody is associated with immune complex–mediated disease after infection. We detected high titers of serum antibody of low avidity for H1-2009 antigen, and low-avidity pulmonary immune complexes against the same protein, in severely ill individuals. Moreover, C4d deposition—a marker of complement activation mediated by immune complexes—was present in lung sections of fatal cases. Archived lung sections from middle-aged adults with confirmed fatal influenza 1957 H2N2 infection revealed a similar mechanism of illness. These observations provide a previously unknown biological mechanism for the unusual age distribution of severe cases during influenza pandemics.
Similar content being viewed by others
Main
Pandemic viruses may promote bacterial infections1, injure the lungs2,3,4, decrease type I interferon (IFN) abundance5, promote a cytokine storm and induce apoptosis6 to cause severe disease. Although these are all attractive hypotheses, the explanation for the enhanced severity of cases in middle-aged adults during pandemics7,8,9,10 remains unclear.
In 2009, a new H1N1 influenza A virus caused severe disease in naive middle-aged individuals with preexisting immunity against seasonal strains11,12,13. In contrast to seasonal disease, the elderly were relatively spared, and young children had milder disease than middle-aged subjects11,12,13,14,15,16. Preexisting neutralizing cross-reactive antibodies elicited by an H1N1 virus that circulated before 1957 protected the elderly11,12. Middle-aged adults had been exposed repeatedly to seasonal influenza viruses, leading to antibody production, whereas young children often lacked previous exposures12. One reason for the severe illness observed in middle-aged individuals could be that an antibody repertoire shaped by seasonal infections may recognize but fail to neutralize the new pandemic strain, leading to immune complex–mediated disease17,18. Here we characterized the pathogenesis of severe pandemic respiratory disease in middle-aged adults with no preexisting comorbidities.
We obtained tracheal and nasopharyngeal aspirates and serum samples reflecting various disease severities in adult outpatients (n = 21) and inpatients (n = 54) infected with 2009 H1N1. The median age of the subjects was 39 years (range 17–57 years). Twenty-three subjects died, 16 (69%) of refractory hypoxemia. Fifteen survivors required intensive care.
In addition, we analyzed nasopharyngeal secretions from adults hospitalized with seasonal influenza A viruses (2007/08) and from infants and young children infected with 2009 H1N1. In Argentina, universal immunization against influenza in children was not recommended until 2010.
Lung sections of individuals with fatal 2009 H1N1 showed widened interalveolar septa, interstitial hemorrhages, abundant intra-alveolar edema with deposition of hyaline membranes and an infiltrate of mononuclear cells (Fig. 1a)10,19. Lungs showed hyperplasia and detachment of type II pneumocytes into the lumen (Fig. 1a). Fatal cases of seasonal H1N1 influenza also showed interstitial edema, desquamation of type II pneumocytes and mononuclear cell infiltration (Fig. 1a). We detected influenza A 2009 H1N1 mainly in epithelial cells of bronchioles; we occasionally detected seasonal H1N1 in respiratory epithelial cells from preexposed elders (Fig. 1b).
(a) Pulmonary histopathology in representative lung sections of fatal 2009 H1N1 and seasonal H1N1 virus-infected humans (H&E). (b) Detection of 2009 H1N1 and seasonal H1N1 influenza viruses. Scale bar, 100 μm. The boxes are details of pulmonary edema (2009 H1N1, H&E), peribronchiolar mononuclear cell infiltration (seasonal H1N1, H&E), and virus-infected cells (anti-H1N1 stains). Scale bars, 100 μm. (c) 2009 H1N1 RT-PCR Ct values in nasopharyngeal secretions of fatal, intensive care unit (ICU) and ambulatory cases; there were no significant differences. (d) 2009 H1N1 RT-PCR Ct values by time of symptoms before nasopharyngeal sampling.
2009 H1N1 RNA (vRNA) expression was similar in outpatients and inpatients requiring intensive care or not surviving (Fig. 1c; P = 0.9). However, vRNA levels correlated with the number of days that symptoms were apparent before consultation (P = 0.027; Fig. 1d), and individuals with severe disease (those who received intensive care or died) were sampled later than outpatients (median 6 d versus 3 d; P = 0.02). Adjusting the relationship between vRNA levels and severity for the number of days that symptoms were apparent did not reach statistical significance (P = 0.3).
Analysis of type I IFN production showed lower tracheal than nasopharyngeal amounts of IFN-α, with similar amounts in 2009 H1N1 as compared to seasonal influenza infections (Supplementary Fig. 1). IFN-β production was universally low (P = 0.6 pandemic versus seasonal; Supplementary Fig. 1).
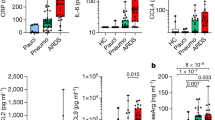
Certain pandemic hemagglutinins are thought to cause a cytokine storm20. Nasopharyngeal secretions in 2009 H1N1 and seasonal infections showed similar amounts of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), IL-1β, IL-10 and IL-12 (Fig. 2 and Supplementary Fig. 2). IL-8 amounts were higher in individuals with pandemic H1N1 influenza (P = 0.01; Fig. 2a).
(a–d) IL-8 (a), TNF-α (b), IL-6 (c) and IL-1β (d) responses in respiratory secretions of individuals infected with 2009 H1N1 influenza virus (TA, tracheal aspirates; NP, nasopharyngeal secretions) or seasonal influenza viruses (H1N1 and H3N2). Comparisons for nasopharyngeal secretions: IL-8, P = 0.01; all other cytokines, not significant. (e–h) IL-1β (e), IL-6 (f), IL-10 (g) and TNF-α (h) production by human monocytes incubated with a dose range of recombinant protein H1-1918, H1-1999, H1-2009, avian H5, hMPV F and a control monoclonal human IgG against the Sa antigenic site of influenza hemagglutinin (2D1). Representative of three independent experiments. (i) Individual TLR activation in HEK293 cells. Dose: 5 μg of H1-1918, H1-1999, H1-2009, hMPV F and no ligand. Representative of two independent experiments. Error bars represent s.e.m.
Production of inflammatory cytokines by human monocytes isolated from peripheral blood of healthy donors and exposed to H1-2009, H1-1918 and H1-1999 proteins was similar (Fig. 2e–h). Higher amounts of inflammatory cytokines were detected with avian H5 (refs. 21,22). Unexpectedly, the inflammatory response elicited by human monocytes against the human metapneumovirus fusion protein (hMPV F) was higher than against influenza hemagglutinins. In fact, hMPV F was a vigorous Toll-like receptor 2 (TLR2) and TLR4 agonist, whereas H1 2009 and H1-1918 were weak TLR2 and TLR4 agonists, respectively (Fig. 2i). Seasonal H1-1999 activated TLR4 (Fig. 2i). No hemagglutinin protein activated other TLRs (Fig. 2i).
Individuals with severe pandemic influenza had profound lymphopenia (Fig. 3a)10. Both CD4+T lymphocyte and CD8+T lymphocyte counts were below normal ranges (Fig. 3b,c). Lymphopenia was associated with a lung T lymphocytosis (Fig. 3d), probably explained by the pandemic virus's conservation of numerous T cell epitopes from seasonal strains23,24. In fact, we observed many CD8+ T lymphocytes in lung sections of individuals who had pandemic influenza (Fig. 3e)19. We did not detect significant T lymphocyte apoptosis (Supplementary Fig. 3).
(a–c) CD3+ (a), CD4+ (b) and CD8+ (c) T lymphocyte counts in fatal and ICU cases of 2009 H1N1 influenza A virus on admission. (d,e) Immunohistochemistry for CD3+ T lymphocytes (red arrows) (d), CD8+ T lymphocytes (yellow arrows) (e) in representative lung sections of fatal influenza 2009 H1N1 and seasonal infection. Positive controls are archived sections from different organs in unrelated subjects provided for every antibody. Negative controls are lung sections from an individual who died from a nonpulmonary disease (scale bars, 100 μm).
We then asked whether lung lymphocytosis was associated with T helper type 2 (TH2) immunopathogenesis, a mechanism of immune-mediated viral respiratory illnesses25. Analysis of IFN-γ (TH1), IL-4 (TH2) and IL-17 (TH17) abundance showed few cytokine-positive cells in lung sections and low cytokine concentrations in secretions from individuals with pandemic H1N1 influenza (data not shown).
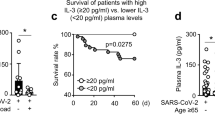
2009 H1N1 virus shares 17% of its B cell epitopes against hemagglutinin and neuraminidase with seasonal influenza A viruses24. Therefore, we examined whether cross-reactive, nonprotective antibodies against 2009 H1N1 were present in sera of naive adults (Fig. 4a). IgG against hemagglutinin antigens was absent in infants but detected in naive middle-aged adults and in the elderly (Fig. 4a). However, antibody avidity for H1 2009 was lower in middle-aged adults than in elderly individuals (P < 0.05; Fig. 4b). In fact, middle-aged adults had higher avidity for H1-1999 than for H1-2009 (P = 0.03; Supplementary Fig. 4). Moreover, unlike elderly subjects, middle-aged adults lacked protective titers of neutralizing antibody against 2009 H1N1 (Fig. 4c)11,12.
(a) Serum IgG endpoint titers against hemagglutinin proteins by immunoassay in infants (mean age (range), 7.6 months (6.1–11.8); n = 10), middle-aged adults (n = 16) and elderly (n = 12). (b) Protein-specific avidity of IgG after 6–9 M urea wash in naive adults (age range, 25–43 years old) and elderly (age range, 75–97 years old); P < 0.05. (c) Microneutralization titers for infants, adults and the elderly; P < 0.05 for elderly versus both groups. (d) Endpoint titer of serum IgG against H1-2009 antigens by immunoassay in adults with severe (n = 12) versus mild (n = 11) disease, P < 0.05. (e) H1-2009–specific avidity of IgG after 7–9 M urea wash in infected adults with severe (n = 12) versus mild (n = 11) pandemic disease, P < 0.05. (f) H1-2009–specific avidity of IgG from immune complexes after 8 M urea wash in infected adults with severe (n = 14) versus mild (n = 11) pandemic disease, P < 0.05. (g) C4d detection in representative slides from lung sections of two of six middle-aged fatal cases of 2009 H1N1 influenza showing extensive peribronchiolar immune complex–mediated complement activation. There is trace C4d deposition in representative lung section of fatal seasonal H1N1 virus from an elderly woman. Positive control, C4d deposition in a kidney from an adult with immune complex–mediated glomerulonephritis; negative control, a lung section from an adult with solid tumor (scale bars, 100 μm). (h) Serum complement C3 concentrations in adults who required intensive care or hospital ward admission, and infants infected with H1N1 2009 influenza A virus, adults infected with seasonal influenza virus and adults with pulmonary diseases other than influenza in the ICU and floor; P = 0.036. (i) C4d detection in representative slides from lung sections of two fatal 1957 H2N2 influenza cases showing extensive peribronchiolar deposition of complement cleavage product. Negative control from an archived sample with no infectious pulmonary disease. The box in the microphotograph is a detail of lung complement deposition.
IgG against H1-2009 and H1-1999 was present in adults <10 d after 2009 H1N1 infection (Supplementary Fig. 5). Of note, H1-2009–specific IgG titers were higher in severely ill adults as compared to mildly ill adults (Fig. 4d; P = 0.02). Moreover, IgG avidity remained lower for H1 2009 than for H1 1999 in individuals with pandemic H1N1 influenza (P = 0.03; Supplementary Fig. 5), and severely ill individuals had antibody of lower avidity for H1 2009 than mildly ill outpatients (P < 0.05; Fig. 4e). Severe cases also had H1-2009–specific IgG of lower avidity than mild cases in respiratory immune complexes (P < 0.05; Fig. 4f).
Nonprotective antibody responses of low avidity have been associated with immune complex disease in other respiratory infections17,26. We therefore stained lung sections for complement cleavage product C4d27. We detected extensive C4d deposition in bronchioles of individuals infected with 2009 H1N1 (Fig. 4g), matching the distribution in immune complex–mediated diseases due to other viruses26. Conversely, we found trace deposition of C4d in lung sections from individuals infected with seasonal influenza (Fig. 4g).
We detected immune complexes with an anti–complement component C1q assay in secretions of individuals infected with 2009 H1N1 but rarely in samples from individuals infected with seasonal influenza (P = 0.003 comparing floor admissions; Supplementary Fig. 6). Furthermore, we detected higher amounts of immune complexes in secretions of adults admitted with 2009 H1N1 to the intensive care unit compared to those admitted to regular floors (Supplementary Fig. 6).
Confirming our observations, most adults admitted to the intensive care unit with pandemic influenza had low serum C3 concentrations, whereas C3 concentrations were higher in moderately ill subjects (Fig. 4h). Infants infected with 2009 H1N1, adults infected with seasonal influenza and individuals with other pulmonary diseases often had normal C3 concentrations (P = 0.036; Fig. 4h).
Finally, we retrieved archived lung sections from adults who died in Tennessee during the 1957 H2N2 pandemic. Sections of sufficient quality to be processed and stained showed extensive C4d peribronchiolar deposition (Fig. 4i). The presence of influenza A vRNA was confirmed by real time-PCR (data not shown). A control archived lung section from an individual with no pulmonary infection showed no C4d deposition (Fig. 4i).
Taken together, these observations indicate that 2009 H1N1 influenza virus leads to immune complex–mediated disease in adults through high titers of low-avidity nonprotective antibody and immune complex–mediated complement activation in the respiratory tract. Immune complex–mediated lung disease also contributed to fatal cases caused by 1957 H2N2 pandemic influenza. We speculate that this phenomenon contributes to severe symptoms in the middle-aged adult population during all pandemics8.
Young infants and children, as in previous pandemics, had high rates of infection with comparatively low mortality7,11. This paradox is explained by absence of protective and pathogenic immunity in children before infection11,12,13, particularly in countries where pediatric immunization against seasonal viruses is not recommended. Therefore, severe pediatric 2009 H1N1 illness associates with widespread infection in a naive population.
This study addresses several attractive hypotheses advanced to explain the pathogenesis of influenza viruses1,2,3,4,5,6. Although the increased severity of 1918 and 2009 H1N1 pandemic viruses was presumed to be associated with higher pulmonary virus titers2,3,4, a dose-dependent effect on mortality was not described. Similarly, type I IFN modulation seems to play a key part in severe cases of influenza28, but its role in the unusual age distribution of severe cases during pandemic flu remains to be determined.
Secondary bacterial infections were responsible for most deaths in 1918 (ref. 1). During 2009, most fatal cases were primary infections with refractory hypoxemia10,12, and neutrophil lung infiltration was minimal19. Depletion of inflammatory cytokines or pretreatment with steroids did not affect mortality in a mouse model of fatal H5N1 influenza infection29. Moreover, the main neutralizing antigen of milder hMPV elicits substantially more inflammation in vitro than influenza hemagglutinins.
However, other roles for innate immunity may be at work in pathogenesis. For example, Streptococcus pneumoniae nasopharyngeal carriage may also affect illness severity during 2009 H1N1 infection30. Indeed, several factors are likely to contribute to severe pandemic disease in adults and explain the different outcomes in individuals of similar ages and backgrounds.
Certain limitations are inherent to a study of these characteristics. For instance, because we lacked determinations of vRNA levels over time, we cannot disregard a role for viral injury in determining severity. However, the impact of pandemic flu in middle-aged adults compared to infants and elderly argue against a preponderant pathogenic role for direct viral injury, as we would expect a mechanism of illness based on viral virulence to preferentially affect frail populations. Also, we rapidly identified a cross-reactive antibody against 2009 H1N1 known to recognize seasonal H1N1. But whether reactivity against both viruses is identical in stained lung sections is unknown.
In summary, our study provides a new biological explanation for the unusual age distribution of severe cases during pandemic influenza. The association of severe pandemic disease in middle-aged adults with high titers of low-avidity, nonprotective antibody and complement activation by pulmonary immune complexes opens a new paradigm for future therapeutic interventions.
Methods
Subjects and samples.
Archived tracheal aspirates, nasopharyngeal secretions and serum samples were obtained from hospitals in Buenos Aires participating in the National Surveillance Network. 2009 H1N1 infection was confirmed by real-time RT-PCR following the US Centers for Disease Control (CDC) protocol. No samples were obtained from subjects with presumptive (clinical diagnosis) or confirmed (blood, pleural or other sterile site positive culture) bacterial superinfections.
Archived lung sections from cases of fatal primary 2009 H1N1 influenza (n = 9) were also obtained. Seasonal influenza A viruses was confirmed by RT-PCR, culturing or both. The study was approved by the Institutional Review Board of Fundacion INFANT, Hospital Nacional Alejandro Posadas, Hospital Dr. Federico Abete and Vanderbilt University. Informed consent was obtained when appropriate.
Histopathology and immunohistochemistry.
For immunohistochemistry, 4- to 5-micron sections were stained with a polyclonal goat H1N1-specific antibody that cross-reacts with seasonal H1N1 and 2009 H1N1 (Biological Swampscott), CD3-specific antibody (Santa Cruz Biotechnology) and CD8-specific and C4d-specific antibodies (Abcam).
Protein synthesis and purification.
Synthetic, sequence-optimized hemagglutinin constructs were obtained from GeneArt or GenScript and cloned into pcDNA3.1 (Invitrogen). The plasmids were transformed into DH5α Escherichia coli for EndoFree Plasmid Maxi preparation (Qiagen). The DNA was transiently transfected into HEK 293F cells (Invitrogen). The supernatant was harvested on day 7 and purified on nickel columns. Endotoxin contamination was ruled out in hemagglutinin, hMPV F and control proteins after purification with the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (GenScript; sensitivity = 0.005 to 1 European units per ml), according to the manufacturer's instructions.
Immunoassays.
Best-fit titration curves were calculated by nonlinear regression to a sigmoidal function using the GraphPad package (Prism). Endpoint titer was defined as the reciprocal of the highest dilution of sera that had a reading above the cutoff, after subtraction of background in all samples. The cutoff was defined as twice the background signal. Avidity was determined by incubating samples with 6 M, 7 M, 8 M and 9 M urea for 10 min before washing and incubation with secondary antibody.
Type I interferon determinations.
IFN-α, IFN-β and inflammatory cytokines were detected with commercial detection kits (pbl interferon source and BD Biosciences, respectively).
Microneutralization assay.
One hundred half-maximal tissue culture infective dose units of influenza H1N1/A/Arg/17/09 were preincubated with dilutions of serum and then used to infect Madin Darby canine kidney cells in 96-well plates, as previously described12,13. Six replicate wells were used for each antibody dilution. Neutralizing antibody concentrations were determined and were defined as the reciprocal of the highest dilution of serum where 50% of wells were infected, as calculated by the method of Reed and Muench12,13.
Immune complex dissociation and avidity determinations.
Extraction of immune complexes from respiratory secretions was performed by adding 50 μl of 7% polyethylene glycol in PBS to every sample (50 μl). Immune complexes were dissociated by adding 1.5 M glycine hydrochloride (pH 1.8). Control samples were treated with 1.5 M glycine hydrochloride (pH 7.2). Samples were neutralized with 50 μl of 5 mM Tris–hydrochloric acid (pH 7.2). Avidity against H1 2009 was determined as described above.
Monocyte cytokine assays.
Human peripheral blood mononuclear cells were isolated by Ficoll-Hypaque (GE Healthcare Life Sciences) from anonymous healthy donors. Monocytes were isolated with the Monocyte Isolation Kit II (Miltenyi Biotec) as described by the manufacturer. The remaining cells were >95% monocytes, as determined by staining with antibody to CD14 and forward- and side-light scatter analysis using flow cytometry (Becton Dickinson). Purified monocytes (1 × 105 cells per well) were stimulated for 18 h at 37 °C with the respective hemagglutinins in triplicates. Inflammatory cytokines were detected in supernatant fluids of treated and control untreated monocytes with the cytokine bead array kit (BD Biosciences) following the manufacturer's instructions.
Toll-like receptor ligand screening.
TLR agonism was tested by assessing nuclear factor-κB activation in HEK293 cells expressing individual human TLRs (InvivoGen). Hemagglutinins were tested at a dose of 5 μg. All tests were performed in duplicate.
RNA extraction and reverse transcription PCR.
Formalin-fixed, paraffin-embedded autopsy lung tissue specimens were retrieved. Ten-μm sections were deparaffinized and digested, and RNA was extracted with the RecoverAll Total Nucleic Acid Isolation Kit (Ambion) according to the manufacturer's instructions. Extracted RNA was tested by real-time RT-PCR for human RNA and for influenza A and B with the CDC real-time RT-PCR assay to detect seasonal influenza. Matrix gene sequences for 32 1957 H2N2 strains were retrieved from the National Institutes of Health Influenza Virus Resource and aligned with MacVector 11.0 (MacVector). Primer and probe sequences used in the CDC influenza A assay were highly conserved with the 1957 influenza virus sequences (data not shown). Specimens were considered positive for influenza if they had C(t) <60 in two separate reactions.
Statistical analyses.
Data were analyzed using STATA 10.1. Kruskal-Wallis and Mann-Whitney U tests were used where appropriate. Correlation between RNA levels and days of symptoms was explored with the Spearman's rank correlation coefficient test. The impact of RNA levels on disease severity was adjusted by days of flu symptoms using a logistic regression analysis. A P < 0.05 was considered significant.
References
Morens, D.M., Taubenberger, J.K. & Fauci, A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970 (2008).
Tumpey, T.M. et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310, 77–80 (2005).
Itoh, Y. et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460, 1021–1025 (2009).
Kobasa, D. et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323 (2007).
Geiss, G.K. et al. Cellular transcriptional profiling in influenza A virus–infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99, 10736–10741 (2002).
Kash, J.C. et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006).
Taubenberger, J.K. & Morens, D.M. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 12, 15–22 (2006).
Simonsen, L. et al. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J. Infect. Dis. 178, 53–60 (1998).
Morens, D.M. & Fauci, A.S. The 1918 influenza pandemic: insights for the 21st century. J. Infect. Dis. 195, 1018–1028 (2007).
Perez-Padilla, R. et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N. Engl. J. Med. 361, 680–689 (2009).
Chowell, G. et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 361, 674–679 (2009).
Hancock, K. et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361, 1945–1952 (2009).
Centers for Disease Control and Prevention (CDC). Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 58, 521–524 (2009).
The ANZIC Influenza Investigators. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N. Engl. J. Med. 361, 1925–1934 (2009).
Bhat, N. et al. Influenza-associated deaths among children in the United States, 2003–2004. N. Engl. J. Med. 353, 2559–2567 (2005).
Thompson, W.W. et al. Influenza-associated hospitalizations in the United States. J. Am. Med. Assoc. 292, 1333–1340 (2004).
Delgado, M.F. et al. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat. Med. 15, 34–41 (2009).
Reichert, T., Chowell, G., Nishiura, H., Christensen, R.A. & McCullers, J.A. Does glycosylation as a modifier of original antigenic sin explain the case age distribution and unusual toxicity of pandemic novel H1N1 influenza? BMC Infect. Dis. 10, 5 (2010).
Mauad, T. et al. Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Resp. Crit. Care. Med. 181, 72–79 (2010).
Kobasa, D. et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431, 703–707 (2004).
Perrone, L.A., Plowden, J.K., García-Sastre, A., Katz, J.M. & Tumpey, T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4, e1000115 (2008).
de Jong, M.D. et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12, 1203–1207 (2006).
Lewis, D.E., Gilbert, B.E. & Knight, V. Influenza virus infection induces functional alterations in peripheral blood lymphocytes. J. Immunol. 137, 3777–3781 (1986).
Greenbaum, J.A. et al. Preexisting immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. USA 106, 20365–20370 (2009).
Johnson, T.R. et al. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J. Virol. 72, 2871–2880 (1998).
Polack, F.P. et al. A Role for immune complexes in enhanced respiratory syncytial virus disease. J. Exp. Med. 196, 859–865 (2002).
Regele, H. et al. Endothelial C4d deposition is associated with inferior kidney allograft outcome independently of cellular rejection. Nephrol. Dial. Transplant. 16, 2058–2066 (2001).
Hall, C.B., Douglas, R.G., Simons, R.L. & Geiman, J.M. Interferon production in children with respiratory syncytial, influenza and parainfluenza virus infections. J. Pediatr. 93, 28–32 (1978).
Salomon, R., Hoffmann, E. & Webster, R.G. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc. Natl. Acad. Sci. USA 104, 12479–12481 (2007).
Palacios, G. et al. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS ONE 4, e8540 (2009).
Acknowledgements
Funded by the Fundacion INFANT 2008 Fundraising Campaign and AI-054952 (F.P.P.), the Thrasher Research Fund Early Career Award and Fogarty International Center International Clinical Research Fellows Program at Vanderbilt (R24 TW007988) (G.A.M. and J.P.B.), US Department of Defense grant HDTRA1-08-10-BRCWMD-BAA and US National Institutes of Health grant P01 AI058113 (J.E.C. Jr.). Doctoral awards from the Consejo Nacional de Investigaciones Cientıficas y Técnicas, Argentina (A.C.M. and J.P.B.).
Author information
Authors and Affiliations
Contributions
F.P.P., G.A.M., K.M.E., J.D.C., J.E.C. Jr., J.V.W., A.C.M., J.P.B. and M.F.L. designed the project. A.C.M., J.P.B., M.F.L., J.Z.H., B.M., L.D., K.P.W., J.V.W., G.A.M. and F.P.P. performed experiments. J.C.K., J.K., J.B., C.R., Le. A., L.D., R.L., V.S., E.B., Li. A., G.C., J.F., L.S., J.J., M.E., J.E.C. Jr. and J.V.W. developed or provided key reagents or contributed samples. F.P.P. supervised the project. A.C.M., J.P.B., M.F.L., K.M.E., J.D.C., G.A.M. and F.P.P. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 633 kb)
Rights and permissions
About this article
Cite this article
Monsalvo, A., Batalle, J., Lopez, M. et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat Med 17, 195–199 (2011). https://doi.org/10.1038/nm.2262
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2262
This article is cited by
-
A murine monoclonal antibody against H5N1 avian influenza virus cross-reacts with human kidney cortex cells
Archives of Microbiology (2023)
-
Low quality antibody responses in critically ill patients hospitalized with pandemic influenza A(H1N1)pdm09 virus infection
Scientific Reports (2022)
-
Potential impact of individual exposure histories to endemic human coronaviruses on age-dependent severity of COVID-19
BMC Medicine (2021)
-
Persistently high antibody responses after AS03-adjuvanted H1N1pdm09 vaccine: Dissecting the HA specific antibody response
npj Vaccines (2021)
-
Immunologic mechanisms of seasonal influenza vaccination administered by microneedle patch from a randomized phase I trial
npj Vaccines (2021)