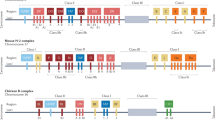
Abstract
The proteasome is key in the cascade of proteolytic processing required for the generation of peptides presented at the cell surface to cytotoxic T lymphocytes by major histocompatibility complex class I molecules. Proteasome-dependent epitope processing is greatly improved through the interferon-γ-induced formation of immunoproteasomes and the activator complex PA28. Tripeptidyl aminopeptidase II also has a strong effect on epitope generation. With its endoproteolytic and exoproteolytic activities, TPPII acts 'downstream' of the proteasome and relies on products released by the proteasome. The antigen-processing cascade involving different proteolytic systems raises anew the question of how antigenic peptides are generated. We therefore revisit the interferon-γ-induced immune adaptation of the proteasome and attempt to redefine its function in connection with the emerging importance of TPPII.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rock, K.L., York, I.A. & Goldberg, A.L. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 4, 670–677 (2004).
York, L.A., Goldberg, A.L., Mo, X.Y. & Rock, K.L. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol. Rev. 172, 29–48 (1999).
Rock, L.K., York, I.A., Saric, T. & Goldberg, A.L. Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–59 (2002).
Falk, K., Rötschke, O., Stevanovic, S., Jung, G. & Rammensee, H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991).
Ortmann, B. et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science 277, 1306–1309 (1997).
Kloetzel, P.M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell. Biol. 2, 179–187 (2001).
Beninga, J., Rock, K.L. & Goldberg, A.L. Interferon-γ can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 273, 18734–18742 (1998).
Stoltze, L. et al. Two new proteases in the MHC class I processing pathway. Nat. Immunol. 1, 413–418 (2000).
York, I.A. et al. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity 3, 429–440 (2003).
Saric, T. et al. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J. Biol. Chem. 276, 36474–36481 (2001).
Geier, E. et al. A giant protease with potential to substitute for some functions of the proteasome. Science 283, 978–981 (1999).
Seifert, U.V. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat. Immunol. 4, 375–379 (2003).
Serwold, T., Gonzalez, F., Kim, J., Jacob, R. & Shastri, N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 (2002).
Saric, T. et al. An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 12, 1169–1176 (2002).
Tanioka, T. et al. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamiliy of aminopeptidases. J. Biol. Chem. 278, 32275–32283 (2003).
Fruci, D., Niedermann, G., Butler, R.H. & van Endert, P. Efficient MHC class I-independent amino-terminal trimming of epitope precursor peptides in the endoplasmic reticulum. Immunity 15, 467–476 (2001).
Glickman, M.H. & Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 (1998).
Voges, D., Zwickl, P. & Baumeister, W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 (1999).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–741 (1997).
Groll, M. et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 (2000).
Orlowski, M., Cardozo, C. & Michaud, C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry 32, 1563–1572 (1993).
Eleuteri, A.M., Kohanski, R.A., Cardozo, C. & Orlowski, M. Bovine spleen multicatalytic proteinase complex (proteasome): replacement of X, Y, and Z subunits by LMP7, LMP2 and MECL1 and changes in properties and specificity. J. Biol. Chem. 272, 11824–11831 (1997).
Kisselev, A.F. et al. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 278, 35869–35877 (2003).
Nussbaum, A.K. et al. Cleavage motifs of the yeast proteasome β subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. USA 95, 12504–12509 (1998).
Köhler, A. et al. The axial channel of the proteasome core particle is gated by the rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7, 1143–1152 (2001).
Knowlton, J.R. et al. Structure of the proteasome activator REGα (PA28α). Nature 390, 639–643 (1997).
Wheatley, D., Grisola, S. & Hernandez-Yago, J. Significance of rapid degradation of newly synthesized proteins in mammalian cells: A working hypothesis. J. Theor. Biol. 98, 283–300 (1982).
Schubert, U. et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 (2000).
Reits, E.A., Vos, J.C., Gromme, M. & Neefjes, J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404, 774–778 (2000).
Princiotta, M.F. et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 3, 343–354 (2003).
Yewdell, J.W., Reits, E. & Neefjes, J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 (2003).
Kisselev, A.F., Akopian, T.N., Woo, K.M. & Goldberg, A.F. The sizes of peptides generated from protein by mammalian 26 and 20S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 274, 3363–3371 (1999).
Rock, K.L. et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 (1994).
Cerundolo, V. et al. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and murine cells. Eur. J. Immunol. 27, 336–341 (1997).
Sijts, A. et al. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J. Immunol. 164, 4500–4560 (2000).
Eggers, M. et al. The cleavage preference of the proteasome governs the yield of antigenic peptides. J. Exp. Med. 182, 1865–1870 (1995).
Benham, A.M., Gromme, M. & Neefjes, J. Allelic differences in the relationship between proteasome activity and MHC class I peptide loading. J. Immunol. 161, 83–89 (1998).
Choppin, J. et al. Characteristics of HIV-1 Nef regions containing multiple CD8+ T cell epitopes: wealth of HLA-binding motifs and sensitivity to proteasome degradation. J. Immunol. 166, 6164–6169 (2001).
Driscoll, J., Brown, M.G., Finley, D. & Monaco, J.J. MHC linked LMP products specifically alter peptidase activities of the proteasome. Nature 365, 262–264 (1993).
Ortiz-Navarette, V. et al. Subunit of the 20S proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature 353, 662–664 (1991).
Cerundolo, V., Kelly, A., Elliott, T., Trowsdale, J. & Townsend, A. Genes encoded in the major histocompatibility complex affecting the generation of peptides for TAP transport. Eur. J. Immunol. 25, 554–562 (1995).
Glynne, R. et al. A proteasome related gene between the two ABC transporter loci in the class II region of the human MHC. Nature 353, 357–360 (1991).
Cruz, M., Elenich, L.A., Smolarek, T.A., Menon, A.G. & Monaco, J.J. DNA sequence, chromosomal localization, and tissue expression of the mouse proteasome subunit LMP10 (Psmb10) gene. Genomics 45, 618–622 (1997).
Aki, M. et al. Interferon-γ induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 115, 257–269 (1994).
Griffin, T.A. et al. Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)-inducible subunits. J. Exp. Med. 187, 97–104 (1998).
Groettrup, M., Standera, S., Stohwasser, R. & Kloetzel, P.M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. USA 94, 8970–8975 (1997).
Nandi, D., Woodward, E., Ginsburg, D.B. & Monaco, J.J. Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor subunits. EMBO J. 16, 5363–5375 (1997).
Witt, E. et al. Characterization of the newly identified human Ump1 homologue POMP and analysis of LMP7 (β5i) incorporation into 20S proteasomes. J. Mol. Biol. 301, 1–9 (2000).
Burri, L. et al. Identification and characterization of a mammalian protein interacting with 20S proteasome precursors. Proc. Natl. Acad. Sci. USA 97, 10348–10353 (2000).
Dubiel, W., Pratt, G., Ferrell, K. & Rechsteiner, M. Purification of a 11S regulator of the multicatlytic proteinase. J. Biol. Chem. 267, 22377–22369 (1992).
Soza, A. et al. Expression and subcellular localization of mouse 20S proteasome activator complex PA28. FEBS Lett. 413, 27–34 (1997).
Murata, S. et al. Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. EMBO J. 20, 5898–5907 (2001).
Preckel, T. et al. Impaired immunoproteasome assembly and immune response in PA28−/− mice. Science 286, 2162–2165 (1999).
Dick, T.P. et al. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell 86, 253–262 (1996).
Shimbara, N. et al. Double cleavage production of the CTL epitope by proteasomes and PA28: role of the flanking region. Genes Cells 2, 785–800 (1997).
Groettrup, M. et al. A role for the proteasome regulator PA28α in antigen presentation. Nature 381, 166–168 (1996).
Bose, S., Stratford, F.L., Broadfoot, K.I., Mason, G.G. & Rivett, A.J. Phosphorylation of 20S proteasome α subunit C8 (α7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by γ-interferon. Biochem. J. 378, 177–184 (2004).
Gacynska, M., Rock, K.L. & Goldberg, A.L. Gamma interferon and expression of MHC genes regulate peptide hydrolysis. Nature 363, 262–264 (1993).
Toes, R.E. et al. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194, 1–12 (2001).
Arnold, D. et al. Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature 360, 171–174 (1993).
Momburg, F. et al. Proteasome subunits encoded by the major histocompatibility complex are not essential for antigen presentation. Nature 360, 174–177 (1993).
Yewdell, J., Lapham, C., Bacik, I., Spies, T. & Bennink, J. MHC-encoded proteasome subunits LMP2 and LMP7 are not required for efficient antigen presentation. J. Immunol. 152, 1163–1170 (1994).
Sibille, C. et al. LMP2+ proteasomes are required for the presentation of specific antigens to cytotoxic T lymphocytes. Curr. Biol. 5, 923–930 (1995).
Sijts, A. et al. Structural features of immunoproteasomes determine the efficient generation of a viral CTL epitope. J. Exp. Med. 191, 503–513 (2000).
Gileadi, U. et al. Generation of an immunodominant CTL epitope is affected by proteasome subunit composition and stability of the antigenic protein. J. Immunol. 163, 6045–6052 (1999).
van Hall, T. et al. Differential influence on CTL epitope presentation by controlled expression of either proteasome immuno-subunits or PA28. J. Exp. Med. 192, 483–492 (2000).
Schwarz, K. et al. Overexpression of the proteasome subunits LMP2, LMP7 and MECL-1 but not PA28α/β enhances the presentation of an immunodominant lymphocyte choriomeningitis virus T cell epitope. J. Immunol. 165, 768–778 (2000).
Van Kaer, L. et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity 1, 533–541 (1994).
Boes, B. et al. IFN-γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J. Exp. Med. 179, 901–909 (1994).
Kuckelkorn, U. et al. Incorporation of major histocompatibility complex encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-γ. Eur. J. Immunol. 25, 2605–2611 (1995).
Emmerich, N.P. et al. The human 26S and 20S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J. Biol. Chem. 275, 21140–21148 (2000).
Morel, S. et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity 12, 107–117 (2000).
Van den Eynde, B.J. & Morel, S. Differential processing of class I restricted epitope by the standard proteasome and immunoproteasome. Curr. Opin. Immunol. 13, 147–153 (2001).
Sun, Y. et al. Expression of the proteasome activator PA28 rescues the presentation of a cytotoxic T lymphocyte epitope on melanoma cells. Cancer Res. 62, 2875–2882 (2002).
Cascio, P., Hiltton, C., Kisselev, A.F., Rock, L. & Goldberg, A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 20, 2357–2366 (2001).
Peters, B., Janek, K., Kuckelkorn, U. & Holzhütter, H.G. Assessment of proteasomal cleavage probabilities from kinetic analysis of time-dependent product formation. J. Mol. Biol. 318, 847–862 (2002).
Cascio, P., Call, M., Petre, B.M., Walz, T. & Goldberg, A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 21, 2636–2645 (2002).
Knuehl, C. et al. The murine cytomegalovirus pp89 immunodominant H-2Ld epitope is generated and translocated into the endoplasmic reticulum as an 11-mer precursor peptide. J. Immunol. 167, 1515–1521 (2001).
Kahn, S. et al. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 167, 6859–6868 (2001).
Kuckelkorn, U. et al. Link between organ-specific antigen processing by 20S proteasomes and CD8+ T cell-mediated autoimmunity. J. Exp. Med. 195, 983–990 (2002).
Macagno, A. et al. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur. J. Immunol. 29, 4037–4042 (1999).
Kisselev, A.F., Kaganovich, D. & Goldberg, A.L. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the α-rings. J. Biol. Chem. 277, 22260–22670 (2002).
Tanahashi, N. et al. Hybrid proteasomes: Induction by interferon-γ and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275, 14336–14345 (2000).
Yamano, T. et al. Two distinct pathways mediated by PA28 and hsp90 in major histocompatibility complex class I antigen processing. J. Exp. Med. 196, 185–196 (2002).
Li, J. et al. Lysine 188 substitutions convert the pattern of proteasome activation by REGγ to that of REGs α and β. EMBO J. 20, 3359–3369 (2001).
Stohwasser, R., Salzmann, U., Giesebrecht, J., Kloetzel, P.M. & Holzhütter, H.G. Kinetic evidences for facilitation of peptide channelling by the proteasome activator PA28. Eur. J. Biochem. 276, 6221–6229 (2000).
Whitby, F.G. et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–20 (2000).
Kopp, F., Dahlmann, B. & Kuehn, L. Reconstitution of hybrid proteasomes from purified PA700-20 S complexes and PA28α/β activator: ultrastructure and peptidase activities. J. Mol. Biol. 313, 465–471 (2001).
Zaiss, D.M., Standera, S., Holzhuetter, H., Kloetzel, P. & Sijts, A.J. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 457, 333–338 (1999).
McCutchen-Maloney, S.L. et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 275, 18557–18565 (2000).
Zaiss, D., Standera, S., Kloetzel, P.M. & Sijts, A. PI31 is a modulator of proteasome formation and antigen processing. Proc. Natl. Acad. Sci. USA 99, 1169–1176 (2002).
Tomkinson, B., Wernstedt, C., Hellman, U. & Zetterqvist, O. Active site of tripeptidyl peptidase II from human erythrocytes is of the subtilisin type. Proc. Natl. Acad. Sci. USA 84, 7508–7512 (1987).
Tomkinson, B. Tripeptidyl peptidases: enzymes that count. Trends Biochem. Sci. 24, 35–359 (1999).
Lévy, F. et al. The final N-terminal trimming of a subaminoterminal proline-containing HLA class I restricted antigenic peptide in the cytosol is mediated by two peptidases. J. Immunol. 169, 4161–4171 (2002).
Reits, E. et al. A role for TPPII in trimming proteasomal degradation products for MHC Class I antigen presentation. Immunity 20, 495–506 (2004).
Wang, E.W. et al. Integration of the ubiquitin-proteasome pathway with a cytosolic oligopeptidase activity. Proc. Natl. Acad. Sci. USA 97, 9990–9995 (2000).
Rape, M. & Jentsch, S. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4, E113–E116 (2002).
Palombella, V.J., Rando, O.J., Goldberg, A.L. & Maniatis, T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 8, 773–785 (1994).
Zhang, S., Skalsky, Y. & Garfinkel, D.J. MGA2 or SPT23 is required for transcription of the Δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151, 473–483 (1999).
Cardozo, C. & Michaud, C. Proteasome degradation of Tau proteins occurs independently of the chymotrypsin-like activity by a non-processive pathway. Arch. Biochem. Biophys. 408, 103–110 (2002).
Wang, R., Chait, B., Wolf, R., Kohanski, C. & Cardozo, C. Lysozyme degradation by the bovine multicatalytic proteinase complex (proteasome):evidence for a nonprocessive mode of degradation. Biochemistry 38, 14573–14581 (1999).
Reits, E. et al. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity 18, 97–108 (2003).
Acknowledgements
Supported by Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Kloetzel, P. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol 5, 661–669 (2004). https://doi.org/10.1038/ni1090
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1090
This article is cited by
-
A transformer-based model to predict peptide–HLA class I binding and optimize mutated peptides for vaccine design
Nature Machine Intelligence (2022)
-
Bortezomib: a proteasome inhibitor for the treatment of autoimmune diseases
Inflammopharmacology (2021)
-
The correlated expression of immune and energy metabolism related genes in the response to Salmonella enterica serovar Enteritidis inoculation in chicken
BMC Veterinary Research (2020)
-
Proteasome subunit α1 overexpression preferentially drives canonical proteasome biogenesis and enhances stress tolerance in yeast
Scientific Reports (2019)
-
N-Octanoyl-Dopamine inhibits cytokine production in activated T-cells and diminishes MHC-class-II expression as well as adhesion molecules in IFNγ-stimulated endothelial cells
Scientific Reports (2019)