Abstract
DEAD-box (DDX) helicases are vital for the recognition of RNA and metabolism and are critical for the initiation of antiviral innate immunity. Modification of RNA is involved in many biological processes; however, its role in antiviral innate immunity has remained unclear. Here we found that nuclear DDX member DDX46 inhibited the production of type I interferons after viral infection. DDX46 bound Mavs, Traf3 and Traf6 transcripts (which encode signaling molecules involved in antiviral responses) via their conserved CCGGUU element. After viral infection, DDX46 recruited ALKBH5, an 'eraser' of the RNA modification N6-methyladenosine (m6A), via DDX46's DEAD helicase domain to demethylate those m6A-modified antiviral transcripts. It consequently enforced their retention in the nucleus and therefore prevented their translation and inhibited interferon production. DDX46 also suppressed antiviral innate immunity in vivo. Thus, DDX46 inhibits antiviral innate responses by entrapping selected antiviral transcripts in the nucleus by erasing their m6A modification, a modification normally required for export from the nucleus and translation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 September 2017
In the version of this article initially published, the label 'siDDX46' in the key to Figure 5g is incorrect. That label should read 'siALKBH5'. The error has been corrected in the HTML and PDF versions of this article.
References
Schlee, M. & Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580 (2016).
Ivashkiv, L.B. & Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Gack, M.U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Cao, X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016).
Zheng, Q. et al. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 25, 1121–1136 (2015).
Portilho, D.M. et al. Endogenous TRIM5α function is regulated by SUMOylation and nuclear sequestration for efficient innate sensing in dendritic cells. Cell Rep. 14, 355–369 (2016).
Ansari, M.A. et al. Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-b responses. PLoS Pathog. 11, e1005019 (2015).
Kim, T.W. et al. Transcriptional repression of IFN regulatory factor 7 by MYC is critical for type I IFN production in human plasmacytoid dendritic cells. J. Immunol. 197, 3348–3359 (2016).
Loo, Y.M. & Gale, M. Jr. Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011).
Zhang, Z. et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 (2011).
Parvatiyar, K. et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161 (2012).
Zhang, Z., Yuan, B., Lu, N., Facchinetti, V. & Liu, Y.J. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol. 187, 4501–4508 (2011).
Schröder, M., Baran, M. & Bowie, A.G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27, 2147–2157 (2008).
Linder, P. & Jankowsky, E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011).
Calo, E. et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253 (2015).
Hoernes, T.P., Hüttenhofer, A. & Erlacher, M.D. mRNA modifications: dynamic regulators of gene expression? RNA Biol. 13, 760–765 (2016).
Wang, X. & He, C. Dynamic RNA modifications in posttranscriptional regulation. Mol. Cell 56, 5–12 (2014).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71, 3971–3975 (1974).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Alarcón, C.R. et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Zhao, X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419 (2014).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Fustin, J.M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013).
Cordin, O. & Beggs, J.D. RNA helicases in splicing. RNA Biol. 10, 83–95 (2013).
Liang, W.W. & Cheng, S.C. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 29, 81–93 (2015).
Bourgeois, C.F., Mortreux, F. & Auboeuf, D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17, 426–438 (2016).
Putnam, A.A. & Jankowsky, E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta 1829, 884–893 (2013).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Xu, Y.Z. et al. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23, 376–385 (2004).
Hirabayashi, R., Hozumi, S., Higashijima, S. & Kikuchi, Y. Ddx46 is required for multi-lineage differentiation of hematopoietic stem cells in zebrafish. Stem Cells Dev. 22, 2532–2542 (2013).
Hozumi, S. et al. DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish. PLoS One 7, e33675 (2012).
Zarnack, K. et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152, 453–466 (2013).
Weber, M. et al. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13, 336–346 (2013).
Sun, X. et al. Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci. Rep. 5, 12521 (2015).
Chen, L.L. & Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478 (2009).
Boutz, P.L., Bhutkar, A. & Sharp, P.A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29, 63–80 (2015).
Bahar Halpern, K. et al. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662 (2015).
Kufel, J., Bousquet-Antonelli, C., Beggs, J.D. & Tollervey, D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol. Cell. Biol. 24, 9646–9657 (2004).
Li, X. et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 17, 806–815 (2016).
Wang, P. et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014).
Liu, J. et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat. Immunol. 15, 612–622 (2014).
Huppertz, I. et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287 (2014).
Moore, M.J. et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9, 263–293 (2014).
Peritz, T. et al. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1, 577–580 (2006).
Katz, Y., Wang, E.T., Airoldi, E.M. & Burge, C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Acknowledgements
We thank T. Fang, M. Jin and Y. Li for technical assistance; X. Liu, C. Han, S. Xu, Y. Han and T. Chen for discussions; and the SIDANSAI Biotechnology Company (Shanghai, China) for helping to generate Ddx46+/− mice. Supported by the National Key Basic Research Program of China (2013CB530503 and 2012CB518900), the National Natural Science Foundation of China (31390431, 81422037 and 81671564) and the CAMS Innovation Fund for Medical Sciences (2016-12M-1-003).
Author information
Authors and Affiliations
Contributions
Q.Z., J.H., Y.Z., and Z.L. performed the experiments; Q.Z., J.H. and X.C. analyzed data and wrote the paper; and X.C. designed and supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
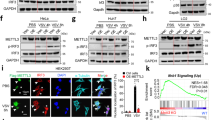
Supplementary Figure 1 DDX46 negatively regulates the production of type I interferons after viral infection.
(a) qRT-PCR analysis of Ifnb mRNA in macrophages transfected with the indicated siRNAs after infected with VSV (MOI=10) as indicated time. (b) Confocal microscopy of macrophages stained with DDX46 antibodies. Scale bar, 10 μm. (c,d) qRT-PCR and immunoblot analysis of DDX46 in macrophages transfected with NC, DDX46 siRNA#1 or #2 as indicated for 72 h. (e) qRT-PCR analysis of Ifnb mRNA expression in human THP1 cells transfected with NC or DDX46 siRNA for 72 h and infected with VSV (MOI=10) for indicated time. (f,g) Immunoblot analysis of the indicated protein in RAW264.7 cell clones stably overexpressing DDX46 (f) or transfected with siRNA for 72 h, and infected with VSV (MOI=10) for indicated time (g). (h,i) qRT-PCR analysis of Ifnb, Il6, Tnf and Il1b mRNA in RAW264.7 cells transfected with NC or DDX46 siRNA for 72 h and infected with VSV (MOI=10) (h) or treated with LPS (100ng/mL) (i) as indicated time. (j) Luciferase activity in HEK293T cells after co-transfection with DDX46 and the indicated plasmids. Luciferase activity is presented relative to Renilla luciferase activity. (k-m) Immunoblot analysis of DDX46 in mouse macrophages infected with VSV (k) or HSV (l) or in mouse peripheral blood mononuclear cells (PBMNC), myeloid conventional dendritic cells (cDC), plasmacytoid dendritic cells (pDC) and in human THP1 cells infected with VSV (m) as indicated time. NS: not significant; **p<0.01, *p<0.05 (Student’s t-test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a,c,e,h -j)) or from three independent experiments with similar results (b,d,f,g,k-m).
Supplementary Figure 2 Gene-ontology (GO) and KEGG analysis of DDX46-bound mRNA with the DAVID tool.
(a) Gel electrophoresis analysis of cDNA for removing free RT primers and size selection. (b) Gel image of amplified cDNA libraries and extraction of the indicated size ranges. (c,d) Overlap of two biological replicates for iCLIP in uninfected or infected RAW264.7 cells. Numbers show the sum of genes identified in each sample. (e) Overlap of transcripts identified by iCLIP of DDX46 in uninfected or infected RAW264.7 cells. Numbers in c,d,e show the sum of genes identified in each sample. (f,g) GO and KEGG analysis of the 1000 most enriched DDX46-bound mRNAs in uninfected cells but not in that of the infected cells. (h,i) GO and KEGG analysis of the 1000 most enriched DDX46-bound mRNAs in infected cell but not in that of uninfected cell. * indicates the enriched antiviral signaling pathway upon viral infection. Data are representative of three independent experiments with similar results (a,b).
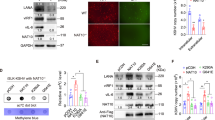
Supplementary Figure 3 DDX46 associates with ALKBH5 to inhibit the expression of antiviral transcripts after viral infection.
(a) Fold enrichment of Gapdh and Ticam1 transcripts in DDX46-RNA co-immunoprecipitation versus RNA-protein input control in RAW264.7 infected with VSV (MOI=10). (b) The relative changes in PSI values observed in 1449 transcripts between wildtype and DDX46+/− macrophages by RNA-seq. (c) RAW264.7 cells were transfected with plasmids for 24 h, and then the cells were fixed, permeabilized, and stained with FISH probes for Tbk1 transcript. Scale bar, 10 μm. (d) RAW264.7 cells were fixed, permeabilized, and stained with FISH probes for Mavs transcript, and then stained with DDX46 and SC-35 antibody. Scale bar, 10 μm. (e) Immunoblot analysis of the indicated protein in macrophages transfected with NC or siRNA for 72 h, and infected with VSV (MOI=10) for indicated time. (f) Immunoblot analysis of the indicated protein in RAW264.7 cells transfected with NC or siRNA for 72 h, and infected with VSV (MOI=10) for indicated time. NS: not significant. (Student’s t-test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a)) or from three independent experiments with similar results (c-f).
Supplementary Figure 4 DDX46 associates with ALKBH5 to inhibit MAVS expression after viral infection.
(a) PAGE gel resolution of immunoprecipitated DDX46 and its associated proteins from peritoneal macrophages infected by VSV for the indicated time. Different bands were analyzed by MS. Arrow indicates the band of the protein detected by MS. (b) IP and immunoblot analysis of HEK293T cells after co-transfection of DDX46 with ALKBH5 upon various stimulation as indicated. (c) Luciferase activity (left) and immunoblot (right) analysis in HEK293T cells after co-transfection with MAVS, SRPK2, and ALKBH5 as indicated. (d) IP and immunoblot analysis of HEK293T cells after co-transfection of DDX46 mutants with ALKBH5 as indicated upon VSV (MOI=10) infection for 6 h. (e,f) Luciferase activity (left) and immunoblot (right) analysis in HEK293T cells after co-transfection with MAVS and DDX46 mutants or truncates as indicated. NS: not significant; *p<0.01 (Student’s t-test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (c(left),e(left), f(left))) or from three independent experiments with similar results (a,b,c(right), d,e(right), f(right)).
Supplementary Figure 5 DDX46-inhibited production od type I interferons is dependent on the recruitment of ALKBH5.
(a,b) qRT-PCR analysis of Ifnb mRNA in RAW264.7 cells transfected with plasmids plus siRNAs as indicated, and infected with VSV (MOI=10) for 12 h. (c) Immunoblot analysis of ALKBH5 in Alkbh5−/− RAW264.7 cells. (d) Fold enrichment of the indicated mRNA transcripts in m6A in vitro IP versus mRNA input control. (e) RAW264.7 cells were fixed, permeabilized, and stained with FISH probes for MAVS transcript, and then stained with DDX46 and ALKBH5 antibody. Scale bar, 10 μm. NS: not significant; *p<0.01 (Student’s t-test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a,b,d)) or from three independent experiments with similar results (c, e).
Supplementary Figure 6 DDX46 negatively regulates the production of inflammatory cytokines and Ifnb in macrophage.
(a) qRT-PCR analysis of Il6, Tnf and Il1b mRNA in peritoneal macrophage cells from Ddx46+/+ or Ddx46+/− mice infected with VSV (MOI=10) as indicated time. (b) qRT-PCR analysis of Ifnb, Il6, Tnf and Il1b mRNA in peritoneal macrophage cells from Ddx46+/+ or Ddx46+/− mice and treated with LPS (100ng/mL) for indicated time. (c) Working model for the mechanism of RNA helicase DDX46 which recruits ALKBH5 to inhibit antiviral innate immunity by nuclear entrapping m6A demethylated antiviral transcripts via m6A modification. Ac: Acetylation. *p<0.05 (Student’s t-test). Data are representative of three independent experiments (mean and s.d. of technical triplicates (a,b)).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–7
Rights and permissions
About this article
Cite this article
Zheng, Q., Hou, J., Zhou, Y. et al. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol 18, 1094–1103 (2017). https://doi.org/10.1038/ni.3830
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3830
This article is cited by
-
Methylated lncRNAs suppress apoptosis of gastric cancer stem cells via the lncRNA–miRNA/protein axis
Cellular & Molecular Biology Letters (2024)
-
Comprehensive analysis of m6A modification in immune infiltration, metabolism and drug resistance in hepatocellular carcinoma
Cancer Cell International (2024)
-
m6A methylation modification and immune infiltration analysis in osteonecrosis of the femoral head
Journal of Orthopaedic Surgery and Research (2024)
-
Interferon-induced transmembrane protein-1 competitively blocks Ephrin receptor A2-mediated Epstein–Barr virus entry into epithelial cells
Nature Microbiology (2024)
-
DDX5 inhibits inflammation by modulating m6A levels of TLR2/4 transcripts during bacterial infection
EMBO Reports (2024)