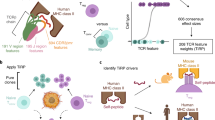
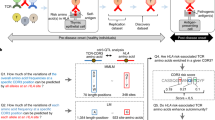
Abstract
Studies of individual T cell antigen receptors (TCRs) have shed some light on structural features that underlie self-reactivity. However, the general rules that can be used to predict whether TCRs are self-reactive have not been fully elucidated. Here we found that the interfacial hydrophobicity of amino acids at positions 6 and 7 of the complementarity-determining region CDR3β robustly promoted the development of self-reactive TCRs. This property was found irrespective of the member of the β-chain variable region (Vβ) family present in the TCR or the length of the CDR3β. An index based on these findings distinguished Vβ2+, Vβ6+ and Vβ8.2+ regulatory T cells from conventional T cells and also distinguished CD4+ T cells selected by the major histocompatibility complex (MHC) class II molecule I-Ag7 (associated with the development of type 1 diabetes in NOD mice) from those selected by a non–autoimmunity-promoting MHC class II molecule I-Ab. Our results provide a means for distinguishing normal T cell repertoires versus autoimmunity-prone T cell repertoires.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Referenced accessions
Protein Data Bank
References
Davis, M.M. & Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Josefowicz, S.Z., Lu, L.F. & Rudensky, A.Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Hogquist, K.A. & Jameson, S.C. The self-obsession of T cells: how TCR signaling thresholds affect fate 'decisions' and effector function. Nat. Immunol. 15, 815–823 (2014).
Vrisekoop, N., Monteiro, J.P., Mandl, J.N. & Germain, R.N. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity 41, 181–190 (2014).
Van Laethem, F. et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 154, 1326–1341 (2013).
Mandl, J.N., Monteiro, J.P., Vrisekoop, N. & Germain, R.N. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013).
Persaud, S.P., Parker, C.R., Lo, W.L., Weber, K.S. & Allen, P.M. Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat. Immunol. 15, 266–274 (2014).
Fulton, R.B. et al. The TCR's sensitivity to self peptide–MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens. Nat. Immunol. 16, 107–117 (2015).
Levine, A.G., Arvey, A., Jin, W. & Rudensky, A.Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014).
Vahl, J.C. et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014).
Trowsdale, J. & Knight, J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 14, 301–323 (2013).
Beringer, D.X. et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 16, 1153–1161 (2015).
Yin, L., Scott-Browne, J., Kappler, J.W., Gapin, L. & Marrack, P. T cells and their eons-old obsession with MHC. Immunol. Rev. 250, 49–60 (2012).
Garcia, K.C. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 33, 429–436 (2012).
Wucherpfennig, K.W., Call, M.J., Deng, L. & Mariuzza, R. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr. Opin. Immunol. 21, 590–595 (2009).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Huseby, E.S. et al. How the T cell repertoire becomes peptide and MHC specific. Cell 122, 247–260 (2005).
Dai, S. et al. Crossreactive T Cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity 28, 324–334 (2008).
Huseby, E.S., Crawford, F., White, J., Marrack, P. & Kappler, J.W. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat. Immunol. 7, 1191–1199 (2006).
Kosmrlj, A., Jha, A.K., Huseby, E.S., Kardar, M. & Chakraborty, A.K. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc. Natl. Acad. Sci. USA 105, 16671–16676 (2008).
Kosmrlj, A. et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465, 350–354 (2010).
Moran, A.E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Garcia, K.C. et al. An αβ T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996).
White, S.H. & Wimley, W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 (1999).
Bouillet, P. et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415, 922–926 (2002).
Zerrahn, J., Held, W. & Raulet, D.H. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell 88, 627–636 (1997).
Merkenschlager, M. et al. How many thymocytes audition for selection? J. Exp. Med. 186, 1149–1158 (1997).
van Meerwijk, J.P. et al. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 185, 377–383 (1997).
Jerne, N.K. The somatic generation of immune recognition. Eur. J. Immunol. 1, 1–9 (1971).
Baker, B.M., Gagnon, S.J., Biddison, W.E. & Wiley, D.C. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity 13, 475–484 (2000).
Chandler, D. Interfaces and the driving force of hydrophobic assembly. Nature 437, 640–647 (2005).
Bogan, A.A. & Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998).
Liu, X. et al. T cell receptor CDR3 sequence but not recognition characteristics distinguish autoreactive effector and Foxp3+ regulatory T cells. Immunity 31, 909–920 (2009).
Ysern, X., Li, H. & Mariuzza, R.A. Imperfect interfaces. Nat. Struct. Biol. 5, 412–414 (1998).
Housset, D. & Malissen, B. What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 24, 429–437 (2003).
Feng, D., Bond, C.J., Ely, L.K., Maynard, J. & Garcia, K.C. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat. Immunol. 8, 975–983 (2007).
Scott-Browne, J.P., White, J., Kappler, J.W., Gapin, L. & Marrack, P. Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature 458, 1043–1046 (2009).
Stadinski, B.D. et al. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 35, 694–704 (2011).
Kanagawa, O., Martin, S.M., Vaupel, B.A., Carrasco-Marin, E. & Unanue, E.R. Autoreactivity of T cells from nonobese diabetic mice: an I-Ag7-dependent reaction. Proc. Natl. Acad. Sci. USA 95, 1721–1724 (1998).
Ridgway, W.M., Ito, H., Fassò, M., Yu, C. & Fathman, C.G. Analysis of the role of variation of major histocompatibility complex class II expression on nonobese diabetic (NOD) peripheral T cell response. J. Exp. Med. 188, 2267–2275 (1998).
Kishimoto, H. & Sprent, J. A defect in central tolerance in NOD mice. Nat. Immunol. 2, 1025–1031 (2001).
Liston, A. et al. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity 21, 817–830 (2004).
Mingueneau, M., Jiang, W., Feuerer, M., Mathis, D. & Benoist, C. Thymic negative selection is functional in NOD mice. J. Exp. Med. 209, 623–637 (2012).
Suri, A., Levisetti, M.G. & Unanue, E.R. Do the peptide-binding properties of diabetogenic class II molecules explain autoreactivity? Curr. Opin. Immunol. 20, 105–110 (2008).
Candéias, S., Katz, J., Benoist, C., Mathis, D. & Haskins, K. Islet-specific T-cell clones from nonobese diabetic mice express heterogeneous T-cell receptors. Proc. Natl. Acad. Sci. USA 88, 6167–6170 (1991).
Lennon, G.P. et al. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity 31, 643–653 (2009).
Todd, J.A. Etiology of type 1 diabetes. Immunity 32, 457–467 (2010).
Marson, A., Housley, W.J. & Hafler, D.A. Genetic basis of autoimmunity. J. Clin. Invest. 125, 2234–2241 (2015).
Day, E.B. et al. Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses. Proc. Natl. Acad. Sci. USA 108, 9536–9541 (2011).
Vanguri, V., Govern, C.C., Smith, R. & Huseby, E.S. Viral antigen density and confinement time regulate the reactivity pattern of CD4 T-cell responses to vaccinia virus infection. Proc. Natl. Acad. Sci. USA 110, 288–293 (2013).
Aronesty, E. ea-utils: “Command-line tools for processing biological sequencing data” https://code.google.com/archive/p/ea-utils/ 2011.
Yang, X. et al. TCRklass: a new K-string-based algorithm for human and mouse TCR repertoire characterization. J. Immunol. 194, 446–454 (2015).
Kruschke, J. Doing Bayesian data analysis: A tutorial introduction with R. (Academic Press, 2010).
Pham-Gia, T. Distributions of the ratios of independent beta variables and applications. Commun. Stat. Theory Methods 29, 2693–2715 (2000).
Cover, T.M. & Thomas, J.A. Elements of Information Theory 2nd edn. (John Wiley & Sons, 2006).
Acknowledgements
Supported by the US National Institutes of Health (RO1-DK095077 and U19 AI109858 to E.S.H., and T32 AI 007349 to B.D.S.), the University of Massachusetts Diabetes and Endocrine Research Center (DK32520 for E.S.H.), Marie Curie Research (I.G.T.), Fundacion Barrie (I.G.T.), the National Institute for Health Research Biomedical Research Centre based at Guy's and St Thomas' National Health Service Foundation Trust and King's College London (M.P.) and the Ragon Institute of MGH, MIT and Harvard (A.K.C.).
Author information
Authors and Affiliations
Contributions
B.D.S., K.S., A.K.S., M.P., A.K.C. and E.S.H. conceived of various aspects of the project and designed and interpreted experiments; B.D.S. performed TCR-sequencing experiments; B.D.S. and K.S. performed statistical analyses; B.D.S., K.S., I.G.-T., J.J., K.S. and E.S.H. performed experiments; and B.D.S., K.S., A.K.C. and E.S.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 A β-strand positions consistently places the residues at CDR3β P6 and P7 within human and mouse TCR-peptide-MHC interfaces.
(a) E8 TCR containing Vβ 13*06 carrying with an 11mer CDR3β loop bound to HLA-DR1 + pTPI, PDB code 2IAM (b) HA1.7 TCR containing Vβ 3.1 carrying with a 14mer CDR3β loop bound to HLA-DR1 + pIAV, PDB code 1FYT (c) KB5-C20 TCR containing Vβ 1*01 carrying with an 18mer CDR3β loop bound to H2-Kb + self-peptide, PDB code 1KJ2. CDR3β loop residues are space filled and shown in orange. A 90o rotation shows the location of the internal disulfide bond that forms between Vβ residues Cys21 to Cys89 that connects the Vβ B and F strands, which fixes the positions the β strand.
Supplementary Figure 2 The ability of residues at CDR3β P6 or P7 to regulate reactivity to self peptide–MHC correlates with hydrophobicity, measured by octanol-water partitioning, but not with the weight or surface area of the amino acids, and correlates weakly with hydropathy.
Correlation plots of P6 or P7 amino acid amino acid fold change in self-reactive TCRs and (a) hydrophobicity based on octanol/water partitioning, amino acid molecular weight (b), solvent assessable surface area42 (c), or hydrophobicity as indexed by Kyte and Doolittle43 (d). These data are derived from the natural log of the average fold increase of the twenty amino acids at the CDR3β residues 6 or 7 in Vβ2+, Vβ6+ and Vβ8.2+ thymocyte TCRs recognizing the MHC alleles: I-Ab, I-Ag7, I-Ad, H2-Kb, H2-Db and H2-Kd. Analysis is nonparametric 2 tailed Spearman correlation (r) computed in Graphpad Prism v6.04.
Supplementary Figure 3 A self-reactivity enrichment- and/or depletion-factor threshold of 0.4 balances the accuracy and completeness of the self-reactivity index.
(a) Scatter plot of CDR3β P6/7-doublets expressed on pre-selection CD4+CD8+ Vβ8.2+ thymocytes derived from C57BL/6 MHC-deficient mice, and pre-selection Vβ8.2+ thymocytes that express CD69 and Nur77-gfp following incubation with β2M-/- I-Ab-expressing cells. Shown are data derived from TCRs carrying CDR3β length of 13 amino acids. CDR3β P6/7-doublets that are significantly enriched (red), unchanged (gray) or depleted (blue) in the I-Ab + self-peptide reactive repertoire are shown. (b) Clustering of empirically-derived self-reactivity enrichment factors for CDR3β 6/7-doublets expressed on Vβ 8.2+ T cells recognizing three alleles of murine MHC class I: H2-Kb, H2-Db, H2-Kd, and three alleles of MHC class II: I-Ab, I-Ag7, IAd. Enrichment factors (rows) were clustered using the k-means algorithm for different values of k and an optimal value of k= 4 was determined based on the “gap-statistic” method44. Self-reactivity indexes were generated based on multiplying the enrichment factors of single amino acids at CDR3β P6 and P7 using threshold cutoffs of ln of 0, 0.2/-0.175, 0.4/-0.35, 0.6/-0.525, 0.8/-0.7, 1/-0.875. The indexes were tested for the ability to identify CDR3β P6/7-doublets that are enriched or deplete on self-reactive thymocytes. (c) Average accuracy in correctly identifying significantly enriched or depleted doublets as red or blue. in Vβ2+, Vβ6+, Vβ8+ TCRs. Accuracy is defined as: the number of doublets correctly identified / number of doublets correctly identified + number of false positive doublets. (d) Average percent of identifying significantly enriched/depleted doublets in Vβ2+, Vβ6+, Vβ8+ TCRs. Completeness is defined as: the number of doublets correctly identified / total number of significantly enriched or depleted doublets.
Supplementary Figure 4 A CDR3β P6 × P7 self-reactivity index with a threshold value of 0.4 accurately predicts whether a CDR3β P6-P7 doublet promotes or limits reactivity to self peptide–MHC.
Fold change of differentially expressed of CDR3β P6-P7 doublets that promote (red), are neutral (white) or limit self-pMHC reactivity (blue) expressed by self-reactive Vβ2+, Vβ6+ or Vβ8.2+ thymocytes as compared to the preselection. Analyses is separated by self-pMHC allele being recognized, (a) I-Ab, (b) I-Ag7, (c) I-Ad, (d) H2-Kb, (e) H2-Db and (f) H2-Kd. The analyses are for Vβ2+, Vβ6+ or Vβ8.2+ TCRs carrying CDR3β loops of 13, 14 and 15 amino acids in length. Self-reactivity enrichment thresholds used are, red > ln(0.4), and blue, < ln (-0.35). P values are from a hypergeometric test of the distribution in red or blue doublets against all significantly different doublets.
Supplementary Figure 5 CDR3β P6-P7 doublets that promote reactivity to self peptide–MHC are greater in frequency among CD4+ Treg cells than among CD4+ Tconv cells and lead to increased levels of CD5 in CD4+ Tconv cells.
(a) Relative fold change of differentially expressed of CDR3β P6-P7 doublets that promote (red), are neutral (white) or limit self-pMHC reactivity (blue) expressed by Vβ2+, Vβ6+ or Vβ8.2+ mouse Tregs versus CD4+ Tconv in NOD.H2b, C57BL/6.H2g7, and NOD mice. P-values are derived from a hypergeometric test. (b-d) CD5 expression level on CD4+ Tconv cells (b), on CD8+ T cells (c), and CD4+ Tregs (d) from splenocytes in C57BL/6 plus TCRβ chain transgenic YAe62 FW, B3K506 SS, or retrogenic Vβ8.2 SW, Vβ8.2 AW, Vβ8.2 EG, Vβ8.2 EQ mixed bone-marrow chimeric mice. CD5 ratios are calculated from the TCRβ transgenic containing cells compared to the same non-transgenic C57BL/6 derived cell populations within the same mouse. Splenocyte populations are pre-gated for TCRβ+ B220- and CD4 or CD8 and Foxp3 expression. Data are derived from YAe62β FW (n=5), Vβ8.2 SW (n=8), Vβ8.2 AW (n=6), B3K506 SS (n=4), Vβ8.2 EG (n=5), Vβ8.2 EQ (n=3) mice. Error bars indicate standard deviation. Statistical significance was assessed by one-way ANOVA: ****p < 0.0001.
Supplementary Figure 6 The H-2g7 haplotype is the main contributing factor to the repertoire-wide shifts toward self-reactivity in the CD4+ Tconv cell repertoire of NOD mice.
Relative fold change of differentially expressed of CDR3β P6-P7 doublets that promote (red), are neutral (white) or limit self-pMHC reactivity (blue) expressed by CD4+ Tconv (a) or CD8+ Tconv (b) in NOD v. B6, B6.H-2g7 v. B6, NOD v. NOD.H-2b, NOD.H-2b v. B6 and NOD v. B6.H-2g7. Fold change in doublet distribution classified by the self-reactivity index is reported as natural log (ln). p values are derived from a hypergeometric test of the distribution in red or blue doublets against all enriched doublets.
Supplementary Figure 7 Estimating Neff using CV versus f relationship.
(a) Neff calculation illustrated for pre-selection Vβ8 TCRs (top), B6-CD4 Vβ6 TCRs (middle) and Bim-CD4 Vβ2 TCRs (bottom). Within each row, the three panels correspond to the three different CDR3 lengths. Neff is calculated independently for each case. Each panel plots the CV vs. f relationship on a log-log scale. The amino acids are distributed across 12-20 frequency bins such that each bin had at least 8 doublets. The black dots correspond to the average CV within each bin and the dashed black line depicts the best fit according to Eq. 3. The colored lines indicate theoretical CV vs. f curves for various population sizes (N=103,104,105,106) calculated using Eq. 3. The inferred Neff is indicated on each panel, together with the number of raw read counts (Nreads).
(b) Neff estimation on simulated data – We used simulations to explore how Neff compares with the true population size (when known). We simulated doublet counts by sampling a multinomial distribution based the frequency distribution seen in the B6-CD4 data. This for done for various sample sizes (N (true)) ranging from 500 to 100,000, and for each case we simulated scenarios involving varying number of replicates (n(rep) = 3, 5, 10, 100). For each case, Neff was calculated (y-axis) based on the CV vs. f relationship described by Eq. (3) and illustrated in fig. S9. These simulations show that at n=3 replicates (red line), the computed Neff underestimates the true population size.
Supplementary Figure 8 Bayesian statistical and cooperativity analysis for the identification of differential expression in CDR3β P6-P7 doublet frequencies.
(a) The posterior probability distributions (Eqs. (7) and (9)) for doublet frequencies are illustrated for three doublets (FW, AW, GK) across three TCR populations. In each panel, we plot the replicate-specific distributions (red, blue, magenta) and the average distribution (black). (b) The probability distribution of the enrichment factor (Eq. (12)) for three doublets (FW, QK and GM) for three representative comparisons (Left: IAb-activated vs. pre-selection, Vβ2, 15-mer, Middle: Bim-/- CD8+ T cells vs. B6 CD8+ T cells, Vβ6, 13-mer, Right: B6 CD4+ T cells vs. NOD CD4+ T cells, Vβ6, 13-mer) illustrating significant enrichment, depletion and no change, respectively. The black dashed line corresponds to an enrichment factor of 1.
(c) Enrichment factor distributions for doublet FW: Comparison of FW CDR3β doublet enrichment in H-2 Db activated versus preselection TCRs for the 9 different cases, (rows: Vβ8, Vβ6, Vβ2, and columns: CDR3 lengths of 13, 14, 15 amino acids). 6 out of 9 cases show a significant enrichment, while 3 cases, owing to poor statistical power, cannot be called based on our threshold (P(Enrichment) > 0.95). It is noteworthy that in the three non-significant cases, the distribution shows a clear bias to the right.
(d) Cooperativity between amino acid distributions at CDR3β P6 and P7: Bar graphs show the Mutual information between the frequency distributions at P6 and P7, MI(P6, P7) expressed as the percentage of the Shannon entropy at P6 (Panel d) or P7 (Panel e). Within each panel, the calculation was performed on the pre-selection TCRs and the MHC-activated TCRs separately (left and right set of bars).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 1259 kb)
Supplementary Table 1
Number of pMHC contacts created by CDR3β residues - atoms with 4Å distance (XLSX 18 kb)
Supplementary Table 2
PCR primers and bar codes for mouse and human TCR sequencing (XLSX 13 kb)
Supplementary Table 3
Genotype & Phenotype of murine T cells subsets and number of sorted T cells and Sequences analyzed (XLSX 14 kb)
Supplementary Table 4
Genotype of donors and number of sorted T cells (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Stadinski, B., Shekhar, K., Gómez-Touriño, I. et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol 17, 946–955 (2016). https://doi.org/10.1038/ni.3491
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3491
This article is cited by
-
A Novel Homozygous Germline Mutation in Transferrin Receptor 1 (TfR1) Leads to Combined Immunodeficiency and Provides New Insights into Iron-Immunity Axis
Journal of Clinical Immunology (2024)
-
MHC-II heterozygosity limits type 1 diabetes susceptibility through negative selection
Nature Immunology (2023)
-
I-Ag7 β56/57 polymorphisms regulate non-cognate negative selection to CD4+ T cell orchestrators of type 1 diabetes
Nature Immunology (2023)
-
Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus
Nature Communications (2022)
-
Indispensable epigenetic control of thymic epithelial cell development and function by polycomb repressive complex 2
Nature Communications (2021)