Abstract
The physiological functions of members of the tumor-necrosis factor (TNF) receptor (TNFR)–associated factor (TRAF) family in T cell immunity are not well understood. We found that in the presence of interleukin 6 (IL-6), naive TRAF5-deficient CD4+ T cells showed an enhanced ability to differentiate into the TH17 subset of helper T cells. Accordingly, TH17 cell–associated experimental autoimmune encephalomyelitis (EAE) was greatly exaggerated in Traf5−/− mice. Although it is normally linked with TNFR signaling pathways, TRAF5 constitutively associated with a cytoplasmic region in the signal-transducing receptor gp130 that overlaps with the binding site for the transcription activator STAT3 and suppressed the recruitment and activation of STAT3 in response to IL-6. Our results identify TRAF5 as a negative regulator of the IL-6 receptor signaling pathway that limits the induction of proinflammatory CD4+ T cells that require IL-6 for their development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
04 April 2014
In the version of this article initially published online, the labels "WT" and "KO" in Figure 4b were transposed; the legend for Figure 5a erroneously stated that TH17 cells were differentiated with IL-6-IL-6R and TGF-β for 5 d instead of 3 d; and on page 2 of the article, the word "soluble" was erroneously used to describe the coreceptor CD28. These errors have been corrected for the print, PDF and HTML versions of the article.
References
Zhu, J., Yamane, H. & Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010).
Murphy, K.M. & Reiner, S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2, 933–944 (2002).
Hirano, T. Interleukin 6 in autoimmune and inflammatory diseases: a personal memoir. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 86, 717–730 (2010).
Heinrich, P.C. et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374, 1–20 (2003).
Kishimoto, T. Interleukin-6: from basic science to medicine—40 years in immunology. Annu. Rev. Immunol. 23, 1–21 (2005).
Zhou, L. et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 (2007).
Ivanov, I.I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Weaver, C.T., Hatton, R.D., Mangan, P.R. & Harrington, L.E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 25, 821–852 (2007).
Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V.K. IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 (2009).
Diehl, S. & Rincon, M. The two faces of IL-6 on Th1/Th2 differentiation. Mol. Immunol. 39, 531–536 (2002).
Neurath, M.F. & Finotto, S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 22, 83–89 (2011).
Stritesky, G.L. et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity 34, 39–49 (2011).
Hildebrand, J.M. et al. Roles of tumor necrosis factor receptor associated factor 3 (TRAF3) and TRAF5 in immune cell functions. Immunol. Rev. 244, 55–74 (2011).
Ha, H., Han, D. & Choi, Y. in Current Protocols in Immunololgy (eds. Coligan, J.E., Bierer, B.E., Margulies, D.H., Shevach, E.M. & Strober, W.) Ch. 11, Unit 11.9D, 11.9D.1–11.9D.19 (John Wiley & Sons, 2009).
Häcker, H., Tseng, P.H. & Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11, 457–468 (2011).
Croft, M. Co-stimulatory members of the TNFR family: keys to effective T-cell immunity? Nat. Rev. Immunol. 3, 609–620 (2003).
Sugamura, K., Ishii, N. & Weinberg, A.D. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat. Rev. Immunol. 4, 420–431 (2004).
Watts, T.H. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. 23, 23–68 (2005).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Sun, L., Deng, L., Ea, C.K., Xia, Z.P. & Chen, Z.J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301 (2004).
Xie, P., Kraus, Z.J., Stunz, L.L., Liu, Y. & Bishop, G.A. TNF receptor-associated factor 3 is required for T cell-mediated immunity and TCR/CD28 signaling. J. Immunol. 186, 143–155 (2011).
Tsitsikov, E.N. et al. TRAF1 is a negative regulator of TNF signaling. enhanced TNF signaling in TRAF1-deficient mice. Immunity 15, 647–657 (2001).
Gardam, S., Sierro, F., Basten, A., Mackay, F. & Brink, R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity 28, 391–401 (2008).
King, C.G. et al. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat. Med. 12, 1088–1092 (2006).
Kraus, Z.J., Haring, J.S. & Bishop, G.A. TNF receptor-associated factor 5 is required for optimal T cell expansion and survival in response to infection. J. Immunol. 181, 7800–7809 (2008).
Nakano, H. et al. Targeted disruption of Traf5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc. Natl. Acad. Sci. USA 96, 9803–9808 (1999).
So, T., Salek-Ardakani, S., Nakano, H., Ware, C.F. & Croft, M. TNF receptor-associated factor 5 limits the induction of Th2 immune responses. J. Immunol. 172, 4292–4297 (2004).
Esparza, E.M., Lindsten, T., Stockhausen, J.M. & Arch, R.H. Tumor necrosis factor receptor (TNFR)-associated factor 5 is a critical intermediate of costimulatory signaling pathways triggered by glucocorticoid-induced TNFR in T cells. J. Biol. Chem. 281, 8559–8564 (2006).
Grech, A., Quinn, R., Srinivasan, D., Badoux, X. & Brink, R. Complete structural characterisation of the mammalian and Drosophila TRAF genes: implications for TRAF evolution and the role of RING finger splice variants. Mol. Immunol. 37, 721–734 (2000).
Tada, K. et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J. Biol. Chem. 276, 36530–36534 (2001).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Ye, H., Park, Y.C., Kreishman, M., Kieff, E. & Wu, H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell 4, 321–330 (1999).
Park, Y.C., Burkitt, V., Villa, A.R., Tong, L. & Wu, H. Structural basis for self-association and receptor recognition of human TRAF2. Nature 398, 533–538 (1999).
Arch, R.H. & Thompson, C.B. 4–1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor κB. Mol. Cell. Biol. 18, 558–565 (1998).
Serada, S. et al. IL-6 blockade inhibits the induction of myelin antigen-specific Th17 cells and Th1 cells in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 105, 9041–9046 (2008).
Hirota, K. et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263 (2011).
Bulek, K. et al. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat. Immunol. 12, 844–852 (2011).
Sun, D. et al. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat. Immunol. 12, 853–860 (2011).
Buchta, C.M. & Bishop, G.A. TRAF5 negatively regulates TLR signaling in B lymphocytes. J. Immunol. 192, 145–150 (2014).
Kawamata, S., Hori, T., Imura, A., Takaori-Kondo, A. & Uchiyama, T. Activation of OX40 signal transduction pathways leads to tumor necrosis factor receptor-associated factor (TRAF) 2- and TRAF5-mediated NF-κB activation. J. Biol. Chem. 273, 5808–5814 (1998).
Croft, M., So, T., Duan, W. & Soroosh, P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol. Rev. 229, 173–191 (2009).
Durant, L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010).
Stahl, N. et al. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science 267, 1349–1353 (1995).
Dittrich, E., Haft, C.R., Muys, L., Heinrich, P.C. & Graeve, L. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J. Biol. Chem. 271, 5487–5494 (1996).
Tanaka, Y. et al. c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol. Cell. Biol. 28, 4805–4818 (2008).
Mizushima, S. & Nagata, S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18, 5322 (1990).
Minami, M. et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl. Acad. Sci. USA 93, 3963–3966 (1996).
Kitamura, T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 (2003).
Acknowledgements
We thank W. Heath (University of Melbourne) for OT-II mice; S. Nagata (Kyoto University) and S. Akira (Osaka University) for the Flag-pEF-STAT3 vector. Supported by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (C) (24590571 to T.S.), the Ichiro Kanehara Foundation (T.S.), the Takeda Science Foundation (T.S.), the Suzuken Memorial Foundation (T.S.) and the US National Institutes of Health (AI049453 to M.C.).
Author information
Authors and Affiliations
Contributions
H.Nag., M.C., N.I. and T.S. designed the experiments; H.Nag., Y.O., A.A., T.K., S.Y. and T.S. did the experiments; H.Nag., Y.O., A.A., T.K., S.Y., M.C., N.I. and T.S. analyzed data; H.Nak. contributed reagents and analytical tools; M.C., N.I. and T.S. supervised the project; M.C., N.I. and T.S. wrote the paper; and M.C., N.I. and T.S. provided funding for the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Expression of gp130 and IL-6R protein and Tbx21, Gata3, Foxp3 and Traf5 mRNA in wild-type and Traf5-/- CD4+ T cells.
(a) Expression of IL-6R–gp130 on purified wild-type and Traf5-/- naive CD4+ T cells. (b) Quantitative RT-PCR analysis of the expression of Tbx21, Gata3, and Foxp3 mRNAs in activated CD4+ T cells generated from naive wild-type or Traf5-/- B6 CD4+ T cells cultured for 48 h with anti-CD3 and anti-CD28 in various polarizing conditions (left margin) and presented relative to the expression of the gene encoding β-Actin (average and s.d. of triplicate wells). (c) Quantitative RT-PCR analysis of the expression of Traf5 mRNA in naive and activated wild-type CD4+ T cells cultured for 48 h with anti-CD3 and anti-CD28 in various cytokine conditions (below graph) and presented relative to the expression of the gene encoding β-Actin (average and s.d. of triplicate wells). NS, not significant; **P < 0.01 (Student t-test).
Supplementary Figure 2 Wild-type and Traf5-/- OT-II T cells proliferate and produce IL-6 in response to antigen similarly.
(a,b) Carboxylfluorescein diacetate, succinimidyl ester (CFSE)-dilution in antigen-responding CD4+ T cells generated from naive wild-type or Traf5-/- OT-II CD4+ T cells stimulated for 3 d with wild-type B6 splenic APCs (after depletion of T cells) and indicated concentrations (above lanes) of OVA peptide (amino acids 323-339) (a) or 0.1 μM OVA peptide in the presence of various concentration (above lanes) of IL-6 (b). (c) Primary IL-6 in supernatants of activated CD4+ T cells generated from naive wild-type or Traf5-/- OT-II CD4+ T cells stimulated for 3 d with indicated concentrations of OVA peptide (horizontal axis) and wild-type B6 APCs. (d) TGF-β-mediated proliferation arrest evaluated by CFSE-dilution in antigen-responding CD4+ T cells generated from naive wild-type or Traf5-/- OT-II CD4+ T cells stimulated for 3 d with wild-type B6 APCs and 0.1 μM OVA peptide with or without 10 ng/ml TGF-β.
Supplementary Figure 3 Expression of gp130, STAT3 and phosphorylated STAT3 protein and Il6st and Traf5 mRNA in various cells from wild-type and Traf5-/- B6 mice.
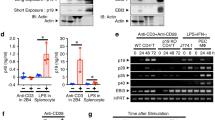
(a) Expression of gp130 (left panel) and immunoblot analysis of phosphorylated STAT3 and total STAT3 after stimulation for various times (above lanes) with 200 ng/ml of IL-6–IL-6R (middle panel) and ratio of phosphorylated STAT3 to total STAT3 after stimulation for 10 min with 200 ng/ml IL-6–IL-6R (average and s.d. of triplicate wells, right panel) in purified splenic wild-type and Traf5-/- polyclonal CD8+ T cells. Isotype, isotype-matched control antibody. (b) Expression of gp130 on T cells, NKT cells, NK cells, B cells, and macrophages from wild-type and Traf5-/- B6 mice. (c) Quantitative RT-PCR analysis of the expression of Traf5 and Il6st mRNAs in different cell populations described above and presented relative to the expression of the gene encoding β-Actin (average and s.d. of triplicate wells). (d) Immunoblot analysis of phosphorylated STAT3 and total STAT3 in wild-type and Traf5-/- macrophages stimulated for 10 min with various concentrations (above lanes) of IL-6–IL-6R (left panel). Ratio of phosphorylated STAT3 to total STAT3 in wild-type and Traf5-/- macrophages stimulated for 10 min with 200 ng/ml of IL-6–IL-6R (average and s.d. of triplicate wells, right panel). (e) Expression of IL-10R (left panel) and immunoblot analysis of phosphorylated STAT3 and total STAT3 (middle panel) and ratio of phosphorylated STAT3 to total STAT3 (average and s.d. of triplicate wells, right panel) after stimulation for 10 min with 50 ng/ml of IL-10 in purified splenic wild-type and Traf5-/- polyclonal CD4+ T cells. (f) Expression of IL-21R (left panel) and immunoblot analysis of phosphorylated STAT3 and total STAT3 (middle panel) and ratio of phosphorylated STAT3 to total STAT3 (average and s.d. of triplicate wells, right panel) after stimulation for 10 min with 10 ng/ml of IL-21 in purified splenic wild-type and Traf5-/- polyclonal CD4+ T cells. NS, not significant; *P < 0.05 and **P < 0.01 (Student t-test).
Supplementary Figure 4 The cytoplasmic amino acid residues in gp130 responsible for TRAF5 binding.
(a) Immunoassay of HEK cells transduced with plasmids encoding gp130 mutants (above lanes) with various deletions in the cytoplasmic region in positions 641–917 (above blot) and cotransfected to express V5-TRAF5 (242–558), followed by immunoprecipitation of proteins from lysates with control IgG or anti-c-Myc and immunoblot analysis with anti-V5 or anti-c-Myc. (b) Amino acid sequence alignment of the TRAF5 binding sites in gp130 from various species. (c) Immunoprecipitation of V5-TRAF5 (242–558) from lysates of HEK cells transiently transfected with plasmid vectors encoding V5-TRAF5 (242–558) and GFP-tagged gp130 (769–800) with wild-type sequence (WT) or alanine substitutions (Ala-mut) (above blot), followed by immunoblot analysis with anti-V5 or anti-GFP. Input (bottom), immunoblot analysis of lysates without immunoprecipitation. (d) Immunoprecipitation of c-Myc-gp130 (1–917) from lysates of HEK cells transiently transfected with plasmid vectors encoding c-Myc-gp130 (1–917), V5-TRAF5 (242–558), and GFP-tagged gp130 (769-800) with wild-type sequence (WT) or alanine substitutions (Ala-mut) (above blot), followed by immunoblot analysis with anti-V5, anti-c-Myc, or anti-GFP.
Supplementary Figure 5 The role of TRAF5 in active and passive EAE.
(a) Body weight changes in wild-type and Traf5-/- mice after induction of active EAE as in Fig. 6a–c, monitored over 22 d (average and s.e.m. of n = 10 mice per genotype). (b) The percentages of donor CD4+CD45.2+ cells in the peripheral blood CD4+ T cells from recipient CD45.1+ B6.SJL mice 7 days after induction of passive EAE as in Fig. 6d (average and s.e.m. of n = 3 mice (no cell transfer) or 6 mice (adoptive transfer)). NS, not significant; *P < 0.05 (Student t-test).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 3275 kb)
Rights and permissions
About this article
Cite this article
Nagashima, H., Okuyama, Y., Asao, A. et al. The adaptor TRAF5 limits the differentiation of inflammatory CD4+ T cells by antagonizing signaling via the receptor for IL-6. Nat Immunol 15, 449–456 (2014). https://doi.org/10.1038/ni.2863
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2863
This article is cited by
-
Identification of necroptosis-related gene TRAF5 as potential target of diagnosing atherosclerosis and assessing its stability
BMC Medical Genomics (2023)
-
The fiber metabolite butyrate reduces gp130 by targeting TRAF5 in colorectal cancer cells
Cancer Cell International (2020)
-
TRAF Molecules in Inflammation and Inflammatory Diseases
Current Pharmacology Reports (2018)
-
Mechanisms and consequences of Jak–STAT signaling in the immune system
Nature Immunology (2017)
-
Oral CD103−CD11b+ classical dendritic cells present sublingual antigen and induce Foxp3+ regulatory T cells in draining lymph nodes
Mucosal Immunology (2017)