Abstract
Genetic studies using mitochondrial DNA (mtDNA) have identified extensive matrilinear diversity among domestic horses. Here, we show that this high degree of polymorphism is not matched by a corresponding patrilinear diversity of the male-specific Y chromosome. In fact, a screening for single-nucleotide polymorphisms (SNPs) in 14.3 kb of noncoding Y chromosome sequence among 52 male horses of 15 different breeds did not identify a single segregation site. These observations are consistent with a strong sex-bias in the domestication process, with few stallions contributing genetically to the domestic horse.
Similar content being viewed by others
Main
The finding of >90 mtDNA haplotypes among domestic horses indicates the incorporation of numerous genetic lineages into the breeding stock1,2,3. These data support domestication from geographically separate areas, which may have occurred through diffusion of the required human expertise or through truly independent domestication processes. But genetic diversity in mtDNA only reflects the maternal contribution to the gene pool. Recently, genetic markers from the male-specific Y chromosome have unveiled the patterns of evolution and migration among modern humans, as manifested by paternal genetic architectures4. Y-chromosome markers will also be informative for addressing the genetic and anthropological processes associated with animal domestication5, as males and females may have been treated differently by early human societies6.
To study the genetic contribution of stallions in horse domestication, we screened for SNPs in 14.3 kb of equine Y-chromosome sequence, divided into 37 fragments (Table 1 and Supplementary Methods online). Because many breeds may derive from a small number of founders or from a limited number of registries in the studbook, bottleneck and founder effects may have reduced the genetic diversity within each single breed. Consequently, we used 52 male horses from 15 different breeds for screening, including divergent European (Ardennais, Connemara, Exmoor, Fjord, Gotland, Icelandic, Shetland, North-Swedish and Thoroughbred) and Asian (Akhal Teké, Arab, Caspian Pony, Khuzestan Arab, Malwari and Thai Pony) breeds. These breeds were selected to represent a wide variety of horses and ponies to cover as much as possible of the gene pool of the domestic horse; most of the breeds have old histories and substantial phenotypic variation.
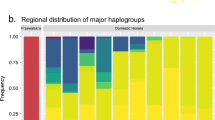
All stallions carried the same Y-chromosome haplotype. This contrasts sharply with the extensive mtDNA diversity and indicates very low levels of Y-chromosome variability in domestic horses. Limited polymorphism is not a general feature of equine nuclear DNA, as a screening of 2.3 kb of X-chromosome intron sequence in a subset of the individuals identified 17 SNPs and a nucleotide diversity (π) of 1.4 × 10−3 ± 0.1 × 10−3 (Supplementary Table 1 online). Nor is it due to selective constraints on the Y chromosome, because, by determining the orthologous X- and Y-chromosome sequence in donkey, we found strong statistical support for lower levels of variability on Y than on X (HKA test P < 0.00001).
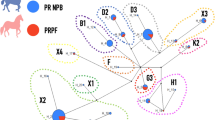
The ancestors to domestic horses have gone extinct in the wild, but a population of Mongolian wild horses, or Przewalski's horses (Equus caballus przewalskii), is kept in captivity. We sequenced one male Przewalski's horse for all Y-chromosome fragments and found that it differed from all domestic horses at six nucleotide positions over the 14.3 kb and also had a 7-bp deletion. Based on Y-chromosome data, the split between Przewalski's horse and domestic horses is estimated to have occurred 120,000–240,000 years ago7. As this is long before wild horses were domesticated (∼6,000 years ago8), wild horses may have had Y-chromosome variability before domestication.
How low is Y-chromosome variability of domestic horses compared with that of, for instance, humans? Several large-scale studies uniformly estimated π in noncoding sequences of the human Y chromosome at 1.0–1.5 × 10−4 (refs. 9–12). One of these studies9 was similar to ours: 50 chromosomes from geographically diverse human samples were screened in 35.3 kb of SMCY intron sequence, identifying 37 segregating sites or one every 950 bp (larger studies found one every 576–840 bp; refs. 9,11,12). A randomization test provided strong statistical support for a significant difference in Y-chromosome polymorphism levels between horses and humans (P < 0.0001). Quantification of this difference is not straightforward, as our domestic horse sample was monomorphic for Y chromosome sequences. But we can conservatively assess the difference by assigning a variable site to the horse data set. With one rare allele at 2% frequency, π would be 3 × 10−6; at 10 % frequency, π would be 1 × 10−5. This implies that π estimates for the Y chromosome are at least 10–30 times lower for domestic horses than for humans.
Our observations are compatible with a scenario of strong sex bias in breeding with only a limited number of sires contributing genetically to the domestic horse (low male effective population size). Modern breeding practice selects stallions and lets them cover many mares each, a breeding scheme that reduces the number of patrilines in the population13. Our data suggest that using a limited number of stallions in breeding may be traditional breeding practice14 and thus date back to the initial phase of horse domestication. If a strong sex bias in breeding were only a modern, breed-specific phenomenon, we would expect to see some Y-chromosome variability among breeds. Moreover, a bias in the early exploitation of each sex could also have contributed to a limited male gene pool. During the Paleolithic age, horses were an important part of the human diet15, and this could have been the case during the early stages of horse domestication as well. In general, food production is maximized if most males are consumed and females are left for reproduction; such a sex bias in exploitation of several domestic animals is suggested from archaeological records6.
Our observations do not exclude the possibility that the equine Y chromosome was low in variability before the time of domestication. The social structure of the wild horses from which domestic breeds evolved probably featured a single stallion holding a harem of multiple mares, implying skewed reproductive success of males. As mtDNA data point at domestication from geographically widespread areas, however, divergent Y-chromosome lineages might have been incorporated into the breeding stock even if local Y-chromosome variability was low. An alternative scenario would imply a single domestication event in a restricted geographical region, resulting in the incorporation of only a limited number of Y-chromosome haplotypes into the breeding stock. When domestic animals, or pastoralism in itself, then spread from one locality to another, the maternal gene pool may have been diversified by the capture of only wild females from local populations (while backcrosses with wild stallions were prevented). Under this scenario, the contrasting levels of variability in mtDNA and the Y chromosome seen in modern horses reflect how the practice of horse domestication spread among early human societies.
Note: Supplementary information is available on the Nature Genetics website.
References
Lister, A.M. et al. Ancient Biomol. 2, 267–280 (1998).
Vilá, C. et al. Science 291, 474–477 (2001).
Jansen, T. et al. Proc. Natl. Acad. Sci. USA 99, 10905–10910 (2002).
Goldstein, D.B. Science 291, 1738–1742 (2001).
MacHugh, D.E. & Bradley, D.G. Proc. Natl. Acad. Sci. USA 98, 5382–5384 (2001).
Zeder, M.A. & Hesse, B. Science 287, 2254–2257 (2000).
Wallner, B. et al. Anim. Genet. 34, 453–456 (2003).
Clutton-Brock, J. A Natural History of Domesticated Mammals. (Cambridge University Press, Cambridge, Massachusetts, 1999).
Shen, P. et al. Proc. Natl. Acad. Sci. USA 97, 7354–7359 (2000).
The International SNP Map Working Group. Nature 409, 928 (2001).
Hammer, M.F. et al. Mol. Biol. Evol. 18, 1189–1203 (2001).
Hammer, M.F. et al. Genetics 164, 1495–1509 (2003).
Cunningham, E.P. et al. Anim. Genet. 32, 360 (2001).
Levine, M.A. J. Anthropol. Archaeol. 18, 29–78 (1999).
Olsen, S.L. Horses Through Time. (Carnegie Museum of Natural History, Pittsburgh, Pennsylvania, 1996).
Acknowledgements
We thank L. Skow for access to the BAC library; M. Webster, J. Seddon and A. Eriksson for help and discussion; and M. Oom, L. Firouz and A. Tawatsin for help with samples. Financial support was obtained from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning. H.E. is a Royal Academy of Sciences Research fellow supported by a grant from the Knut and Alice Wallenberg foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Lindgren, G., Backström, N., Swinburne, J. et al. Limited number of patrilines in horse domestication. Nat Genet 36, 335–336 (2004). https://doi.org/10.1038/ng1326
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1326
This article is cited by
-
Refining the evolutionary tree of the horse Y chromosome
Scientific Reports (2023)
-
Y-chromosome genetic diversity of Bos indicus cattle in close proximity to the centre of domestication
Scientific Reports (2020)
-
Genetic variability and history of a native Finnish horse breed
Genetics Selection Evolution (2019)
-
The horse Y chromosome as an informative marker for tracing sire lines
Scientific Reports (2019)
-
Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes
Nature Communications (2018)